- Carlos Pulido (carlos.pulidoquetglas@unil.ch)
- Daniel Kużnicki (dkuznicki@ibch.poznan.pl)
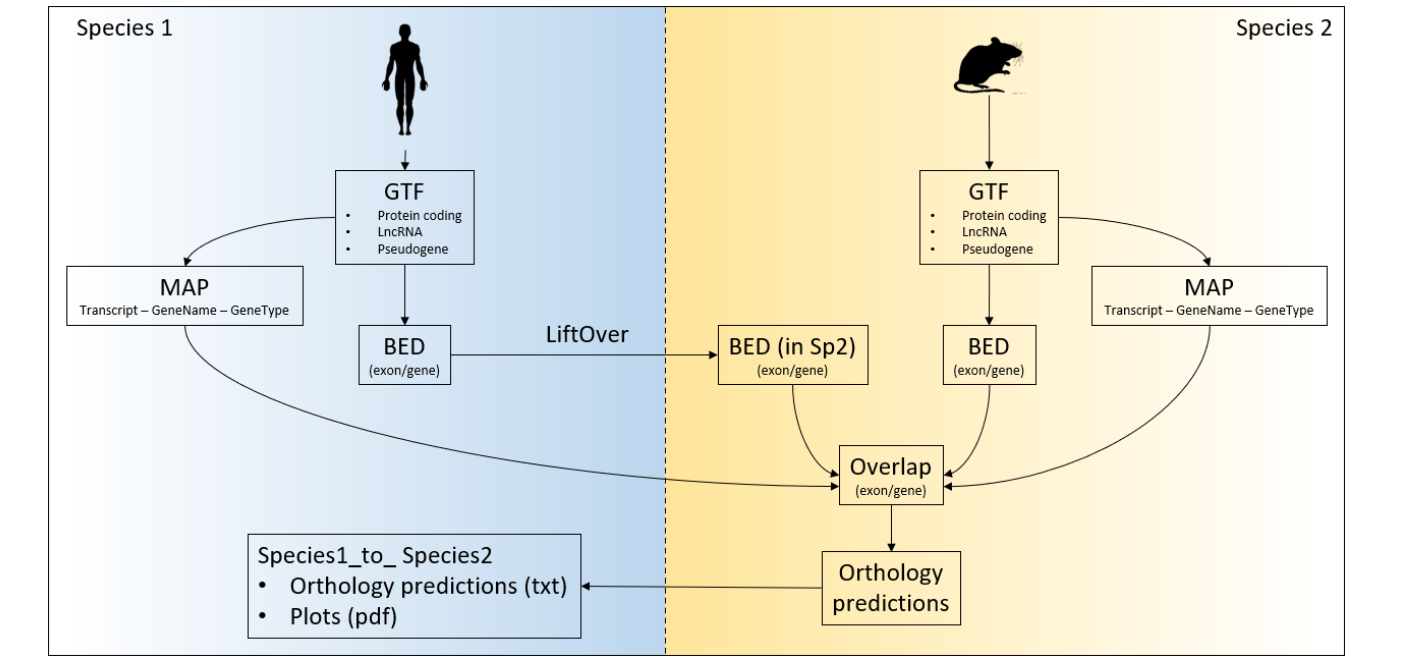
Multiple Species Orthology Finder
Changes in V2:
- Depuration of code
- Simplification and optimization of gene clustering step
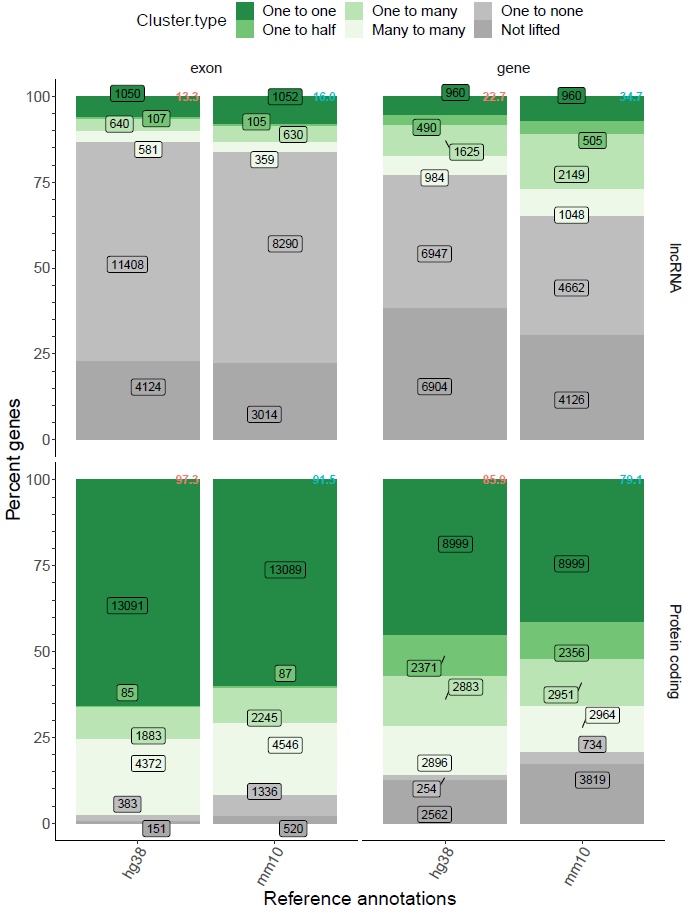
- Enhanced summary plot visualization
- Enhanced summary gene sheet
-
Python3:
- pickle
- pandas
- numpy
- networkx
- matplotlib
- json
- tqdm
-
R:
- ggplot2
- gridExtra
- ggplotify
- dplyr
- scales
- ggrepel
-
BedTools
To execute connector clone this repository locally. Following folders/files must be in the same path from command is executed:
- ConnectOR.py
- config
- dictionaries
- ./scripts/
| species | assembly_version | annotation | chainmap |
|---|---|---|---|
| Human | hg38 | /path/to/human.gtf | |
| Mouse | mm10 | /path/to/mouse.gtf | |
| Zebrafish | danRer10 | /path/to/zebrafish.gtf |
species: name given to each species (must be unique)
assembly_version: ENSEMBL version of the species
annotation [optional]: path to the gtf (recommended to use full path)
chainmap [optional]: comma separated path to each chainmap needed for the analysis (eg for human: "path/to/human_to_mouse.chainmap,path/to/human_to_zebrafish.chainmap"
usage: ConnectOR.v2.py [-h] [-mM MINMATCH] [-g]
ConnectOR (v.2.0) By: Carlos Pulido (carlos.pulido@dbmr.unibe.ch)
optional arguments:
-h, --help show this help message and exit
-mM MINMATCH, --minMatch MINMATCH
0.N Minimum ratio of bases that must remap in liftOver
step.Default: 30 (0.30 minimum ratio)
-g, --gene Generate results at gene level along with exon level
results (default: False)\
Summary table example:
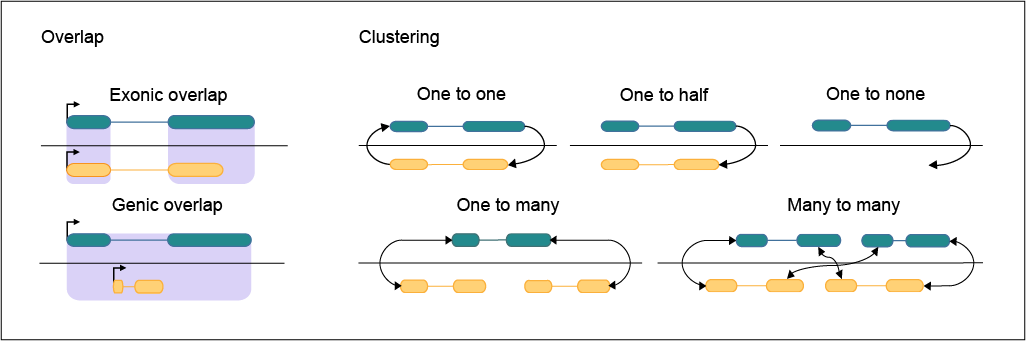
| Gene ID | Species | Biotype | Cluster ID | Cluster Biotype | Cluster type | Gene to hg38 | Gene to mm10 | Orthologues | in_degree | out_degree |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSMUSG00000085871.2 | mm10 | ncRNA | 3434 | ncRNA | One to one | Predicted | ENSG00000223949.8;ROR1-AS1;ncRNA;chr1:0:64171342:- | 1 | 1 | |
| ENSG00000223949.8 | hg38 | ncRNA | 3434 | ncRNA | One to one | Predicted | ENSMUSG00000085871.2;Ube2uos;ncRNA;chr4:0:100384998:- | 1 | 1 |