Welcome to the Human Sinoatrial Node Model repository! This project implements the mathematical model of the human sinus node action potential based on the work by Fabbri et al. [1]. This guide will help you get started with using the model.
The membrane potential equation for the human sinoatrial node cell model is given by:
Here, dV/dt is the rate of change of the membrane potential with respect to time, C is the membrane capacitance, and the terms inside the parentheses represent the various ionic currents that play a vital role in the depolarization and repolarization phases of the action potential.
The currents in this equation, such as the If (Funny current), ICaL (L-type calcium current), ICaT (T-type calcium current), IKr (Rapid delayed rectifier potassium current), IKs (Slow delayed rectifier potassium current), IK,ACh (Potassium current modulated by acetylcholine), Ito (Transient outward potassium current), INa (Sodium current), INaK (Sodium-potassium pump current), INaCa (Sodium-calcium exchanger current), and IKur (Ultra-rapid delayed rectifier potassium current), collectively contribute to the intricate electrical behavior of the sinoatrial node. They are crucial for its proper functioning in generating and conducting cardiac impulses.
To use this model, you'll need:
- Python (version 2.6 or later)
- NumPy library
- JSON library
- SciPy library
- Matplotlib library
- Clone this repository to your local machine:
$ git clone https://github.com/CellularSyntax/HumanSAN-Fabbri2017.git
$ cd HumanSAN-Fabbri2017
- Install the required libraries:
$ pip install numpy scipy matplotlib- Open the
config/config.jsonfile to view and modify the model parameters and initial conditions. - Explore the
tests/test_san_model.jsonfile for an example of how to use theSinoatrialNodeclass to simulate the model and obtain results. - The main implementation of the model is in the
model/SinoatrialNode.pyfile. The classSinoatrialNodecontains methods to initialize the model, update its conditions, calculate constants, and more.
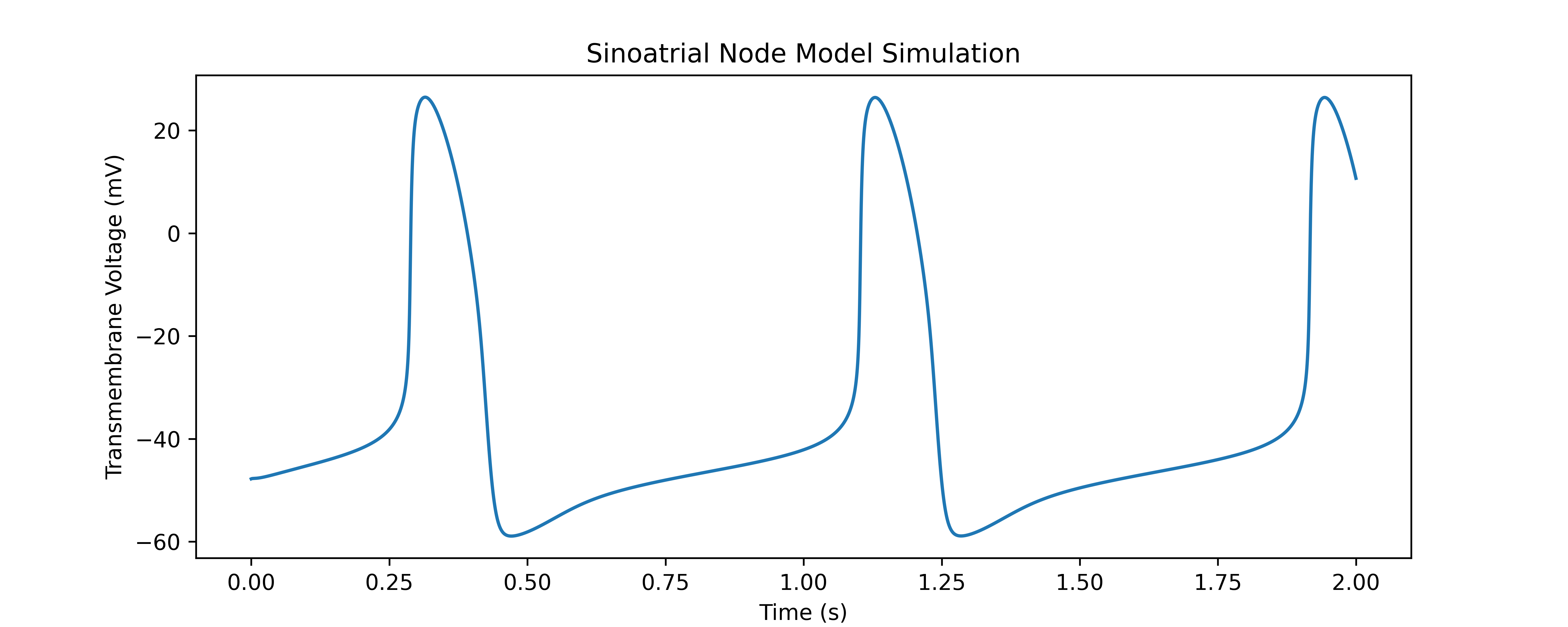
To get started with the Sinoatrial Node Model, you can use the following minimal example to simulate the model and visualize the results using Python:
from model.SinoAtrialNode import SinoAtrialNode
import numpy as np
from scipy.integrate import solve_ivp
from matplotlib import pyplot as plt
from helper_functions import parse_model_parameters
# Load the JSON file containing model parameters
constants, initial_conditions, constant_desc, init_cond_desc = parse_model_parameters("../config/config.json")
# Set the simulation duration and create the SinoAtrialNode object
sim_dur = 2
san = SinoAtrialNode(constant_descriptions=constant_desc,
state_descriptions=init_cond_desc,
constants=constants,
initial_conditions=initial_conditions)
# Solve the model using solve_ivp
sol = solve_ivp(san.calculate_derivatives, [0, sim_dur], list(san.y), method='BDF', rtol=1e-6,
t_eval=np.arange(0, sim_dur, 1e-4), vectorized=False)
# Visualize the results
plt.figure(figsize=(10, 6))
plt.plot(sol.t, sol.y[0], label="Transmembrane Potential")
plt.xlabel("Time (ms)")
plt.ylabel("Voltage (mV)")
plt.title("Sinoatrial Node Model Simulation")
plt.show()The values for constants and initial conditions in the JSON file of that project were obtained from the CellML model available here. These values were derived from the original publication of Fabbri et al. [1].
For a detailed explanation of how this code was implemented and insights into modeling the transmembrane potential, please refer to the corresponding blog article.
If you use this model in your research or projects, please consider citing the original work by Fabbri et al. [1] and acknowledging this repository.
- Fabbri, Alan, et al. "Computational analysis of the human sinus node action potential: model development and effects of mutations." The Journal of Physiology 595.7 (2017): 2365-2396. (DOI: 10.1113/JP273259)