Adapted from the work published in IEEE Transactions on Medical Imaging (https://doi.org/10.1109/TMI.2022.3176598). This project showcases the inference capabilities of transformer-based models for healthcare applications. It has been adapted and developed during the Transformer hackaton @ LabLab.Ai
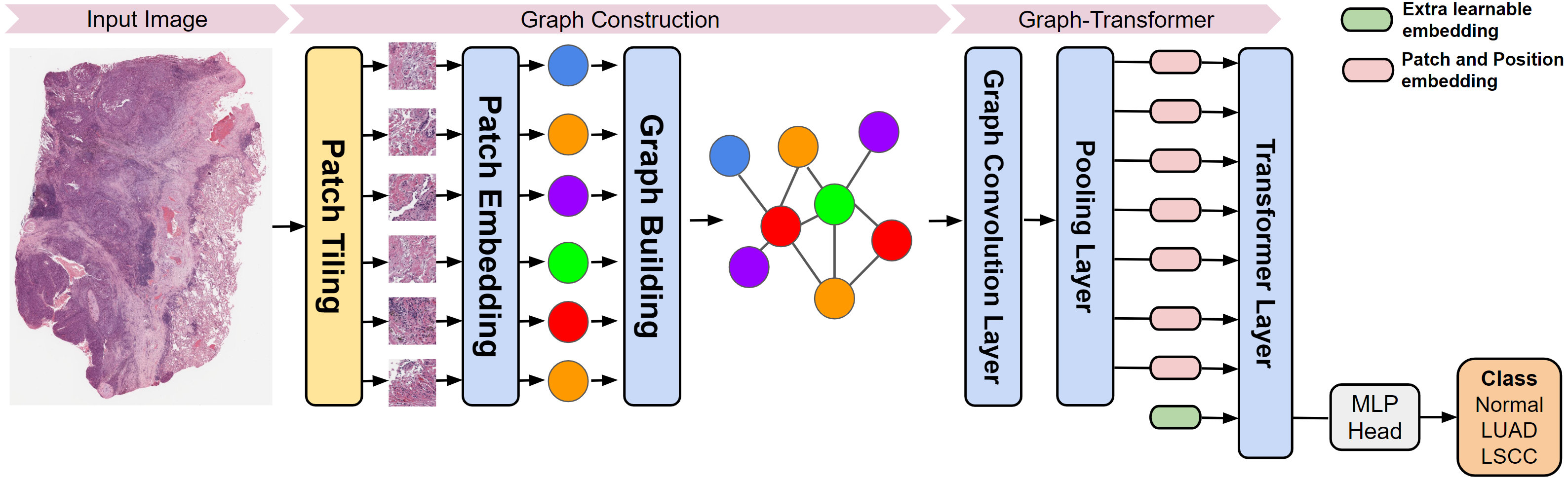
This repository contains a PyTorch implementation of a deep learning based graph-transformer for whole slide image (WSI) classification. The original authors proposed a Graph-Transformer (GT) network that fuses a graph representation of a WSI and a transformer that can generate WSI-level predictions in a computationally efficient fashion.
This adaptation works on the freely available dataset TCGA. The model has been developed considering as primary site the Lungs, for which the available cancer subtypes are LUSC and LUAD. An arbitrary dataset that contains cancer types associated with different primary sites can be built starting from the make_dataset.ipynb notebook. The provided notebook allows creation of datasets of **equally sized WSI slide subsets for cancer subtype**, as well as setting the scale of the whole dataset in gigabytes or terabytes.Start by setting the environment variables by running:
source ./set_env.shNote, for each shell it is necessary to re-run the above command, as it sets the environment variables only for the current shell.
Then, download the required datasets, the gdc tool and the main pip dependencies with:
./download_dataset_manifest.sh
./run_setup.shAt this point, the whole training pipeline can be run by means of:
./run_train_pipeline.shThe above is composed of four main steps:
- extraction of patches from the original slide
- training of the patch feature extractor
- extraction of the graph of each patch obtained in step(1.) for a given slide, by means of the trained feature extractor
- training and testing of the Graph Transformer
Finally, after all training steps are completed, inference can be tested with the streamlit.py demo:
./run_streamlit.shpython src/tile_WSI.py -s 512 -e 0 -j 32 -B 50 -M 20 -o <full_patch_to_output_folder> "full_path_to_input_slides/*/*.svs"
Mandatory parameters:
Go to './feature_extractor' and config 'config.yaml' before training. The trained feature extractor based on contrastive learning is saved in folder './feature_extractor/runs'. We train the model with patches cropped in single magnification (20X). Before training, put paths to all pathces in 'all_patches.csv' file.
python run.py
Go to './feature_extractor' and build graphs from patches:
python build_graphs.py --weights "path_to_pretrained_feature_extractor" --dataset "path_to_patches" --output "../graphs"
Run the following script to train and store the model and logging files under "graph_transformer/saved_models" and "graph_transformer/runs".
bash scripts/train.sh
To evaluate the model. run
bash scripts/test.sh
Split training, validation, and testing dataset and store them in text files as:
sample1 \t label1
sample2 \t label2
LUAD/C3N-00293-23 \t luad
...
To generate GraphCAM of the model on the WSI:
1. bash scripts/get_graphcam.sh
To visualize the GraphCAM:
2. bash scripts/vis_graphcam.sh
Note: Currently we only support generating GraphCAM for one WSI at each time.
More GraphCAM examples:
GraphCAMs generated on WSIs across the runs performed via 5-fold cross validation are shown above. The same set of WSI regions are highlighted by our method across the various cross-validation folds, thus indicating consistency of our technique in highlighting salient regions of interest.