Paper: Di Jin, Mark Heimann, Ryan A. Rossi, Danai Koutra. node2bits: Compact Time- and Attribute-aware Node Representations for User Stitching. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), 2019.
Link: https://gemslab.github.io/papers/jin-2019-node2bits.pdf
Citation (bibtex):
@inproceedings{node2bits-ECML19,
author={Di Jin and Mark Heimann and Ryan A. Rossi and Danai Koutra},
title={Node2BITS: Compact Time- and Attribute-aware Node Representations for User Stitching},
booktitle={ECML/PKDD},
year={2019},
pages={22},
}
Di Jin, Mark Heimann, Ryan A. Rossi, and Danai Koutra. "Node2BITS: Compact Time- and Attribute-aware Node Representations for User Stitching." ECML/PKDD, pp. 22. 2019.
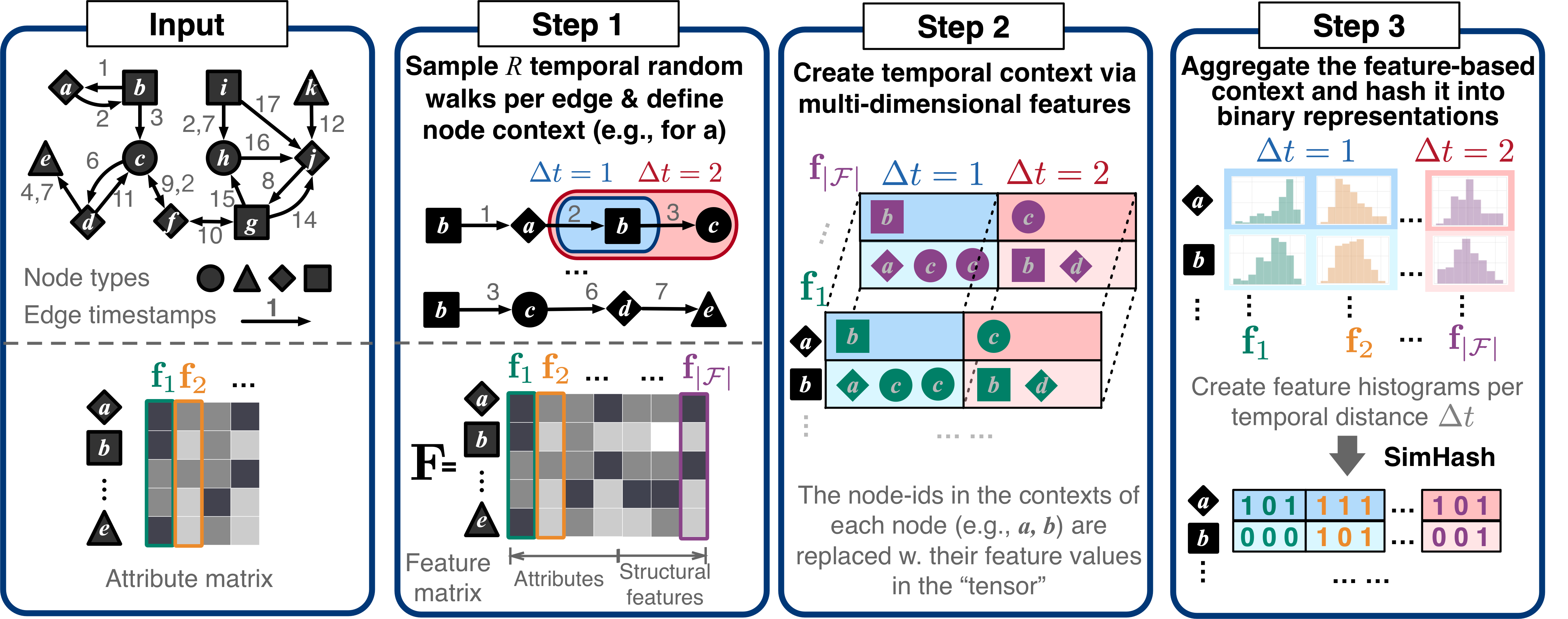
node2bits takes two files as input, the graph file and the category file.
The input graph file can be either static or temporal edge list in the following format separated by tab:
<src> <dst> <weight> <timestamp> (optional)
node2bits will automatically determine if the input graph is static or temporal. The edge list is assumed to be re-ordered consecutively from 0, i.e., the minimum node ID is 0, and the maximum node ID is <#node - 1>. A toy static graph is under "/graph/" directory.
The category file is a mapping between the node ID and its type (e.g., IP, cookie, web agent) with the following format separated by tab:
<category> <id_initial> <id_ending>
if the node IDs are grouped by the type, where <id_initial> and <id_ending> are the starting and ending node ids in type <category>
For example,
0 0 279629
1 279630 283182
means node 0, 1, ... 279629 are in type 0, node 279630, 279631, ... 283182 are in type 1.
But if the node IDs are not grouped by the types, this implementation also supports the following format separated by tab:
<category> <node_id>
which is just the 1-1 mapping. The code accepts either format.
The complete command to run node2bits is as follows.
python main.py --input <graph_file_path> --cat <category_file_path> --output <embedding_file_path> --attri <node_attribute_file_path> --dim <embedding_dimension>
--scope <max_temporal_distance> --base <constant of logarithm binning> --walk_num <#walks_per_node> --walk_length <walk_length>
--walk_mod <temporal_random_walk_bias> --ignore_time <if_ignore_time>
- input, the input graph file stated under the "Graph Input" section above. Default value: '../graph/test.tsv'
- cat, the input category file stated under the "Graph Input" section above. Default value: '../graph/test_cat.tsv'
- attri, the optional input node attribute file. See the exemplar input file for reference: '../graph/test_attri.tsv'. Default value: None
- output, the ouput file of the embedding, which is non-sparse and node-wise binary hashcode. Default value: '../emb/test_emb.txt'
- dim, the dimension of the embedding. Default value: 128
- scope, the maximum temporal distance to consider. Default value: 3
- base, the base constant of logarithm binning. Default value: 4
- walk_num, the number of temporal random walk to perform per node. Default value: 10
- walk_length, the length of the temporal random walk. Default value: 20
- walk_mod, the bias of temporal random walk, can be
<random>, <early>, <late>. Default value: 'early' - ignore_time, a Boolean variable only used when running node2bits on a temporal input graph regardless of its time, i.e., only consider the first 3 columns of the temporal edgelist. Default value: False.
In addition to embedding file indicated in the path output, node2bits also outputs "rep.npz", which is the sparse binary embedding file, and totally <scope> hashtables mapping the bucket_id to node_id in the format:
<bucket_id> [<node_id>]
The hashtables are used to perform unsupervised identity stitching and can be used for AND/OR Amplification.
If you encounter any problems running the code, pls feel free to contact Di Jin (dijin@umich.edu)