-
Notifications
You must be signed in to change notification settings - Fork 49
"HDF5ERROR in aiori-HDF5.c (line 553): cannot create data set" error when benchmarking HDF5 in Blue Gene Q #9
Comments
You are one up on me; IOR with HDF5 won't even compile for me on BG/Q. :) Did you have to make any modifications to get it to build?
Your screenshot says 1.8.8. Are you sure about that?
Sorry, I haven't used that version of IOR in at least two years. Off the top of my head, I know I changed the way IOR gets the host name to identify tasks per node. I think that made IOR's output a little more sane. I cannot remember if anything was more clearly broken than that. |
|
You are right, in my test I used 1.8.8 but I've just tested 1.8.10 and it returns the same error. About the compilation process, see if this helps: I didn't have to modify the source code, just past the parameters into the ./configure and make steps. HDF5 ZLIB Compiler In order to configure it for hdf5 building, I run: Then I just run make normally, and the make command would come up like this: I noticed now that I get two warning in the compilation process: This is the function where the execution dies, maybe its related to this issue? Then when running, I get the following output: HDF5-DIAG: Error detected in HDF5 (1.8.10) MPI-process 0: Began: Tue Jul 16 09:47:10 2013 Test 0 started: Tue Jul 16 09:47:10 2013 access bw(MiB/s) block(KiB) xfer(KiB) open(s) wr/rd(s) close(s) total(s) iter ** error ** Hope this helps |
Yes, I would very much suspect that to be the case. The "(E)" denotes: "Error conditions exist that the compiler can correct, but the program might not produce the expected results." I am a bit surprised that the compiler doesn't halt, actually. I would dig into that error. |
|
Ok thank you. I will start looking into that. Let me know if you need help with the BGQ compilation. |
|
I too am surprised you could build this far. "aiori-HDF5.c", line 555.34: 1506-098 (E) Missing argument(s). means IOR is trying to use old-style HDF5 arguments. There is a backwards compatiblity option, though: add -DH5_USE_16_API to your cflags. Maybe IOR should just do that automatically. I shudder to think anyone is actually running HDF5-1.6.x, but even if they were that define would not affect them. |
|
I guess some day I'll set up my github life so I can "send a pull request. Until then, I blat patches into the issue tracker: |
|
Thanks, Rob! I added that change to master. |
|
It worked!! Thank you for you help Rob. |
Changed the semantics of -R to compare the data with the expected dat…
Hello
I'm having the following error when benchmarking the HDF5 method on a Blue Gene Q: "HDF5ERROR in aiori-HDF5.c (line 553): cannot create data set."
I'm using zlib/1.2.7 and hdf5/1.8.10, and compiling for the compute nodes (not front nodes).
The output is in this attachment below:
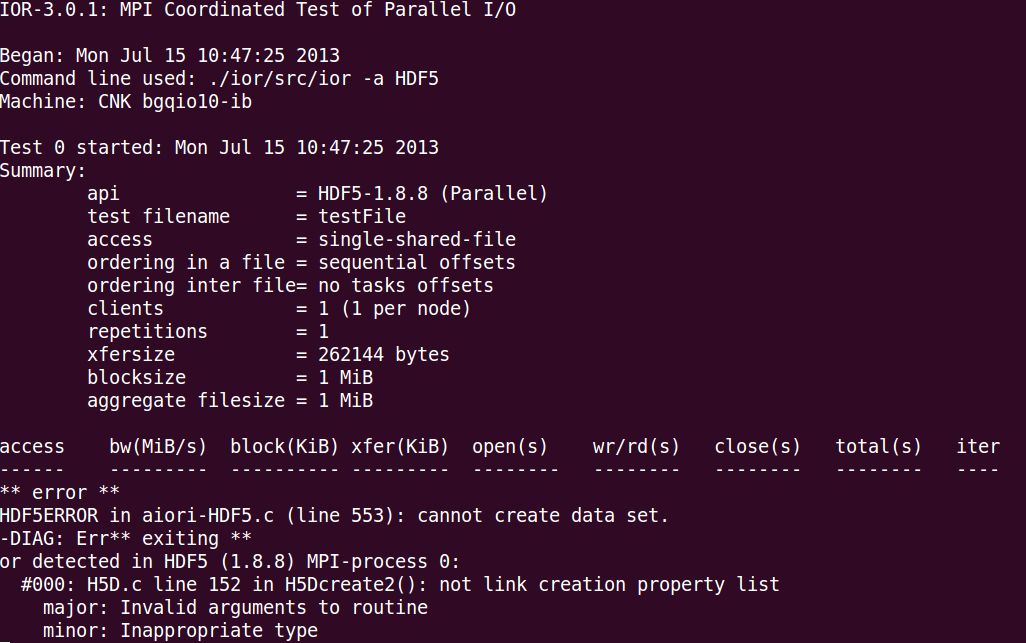
The file creation parameters are in this attachment below:
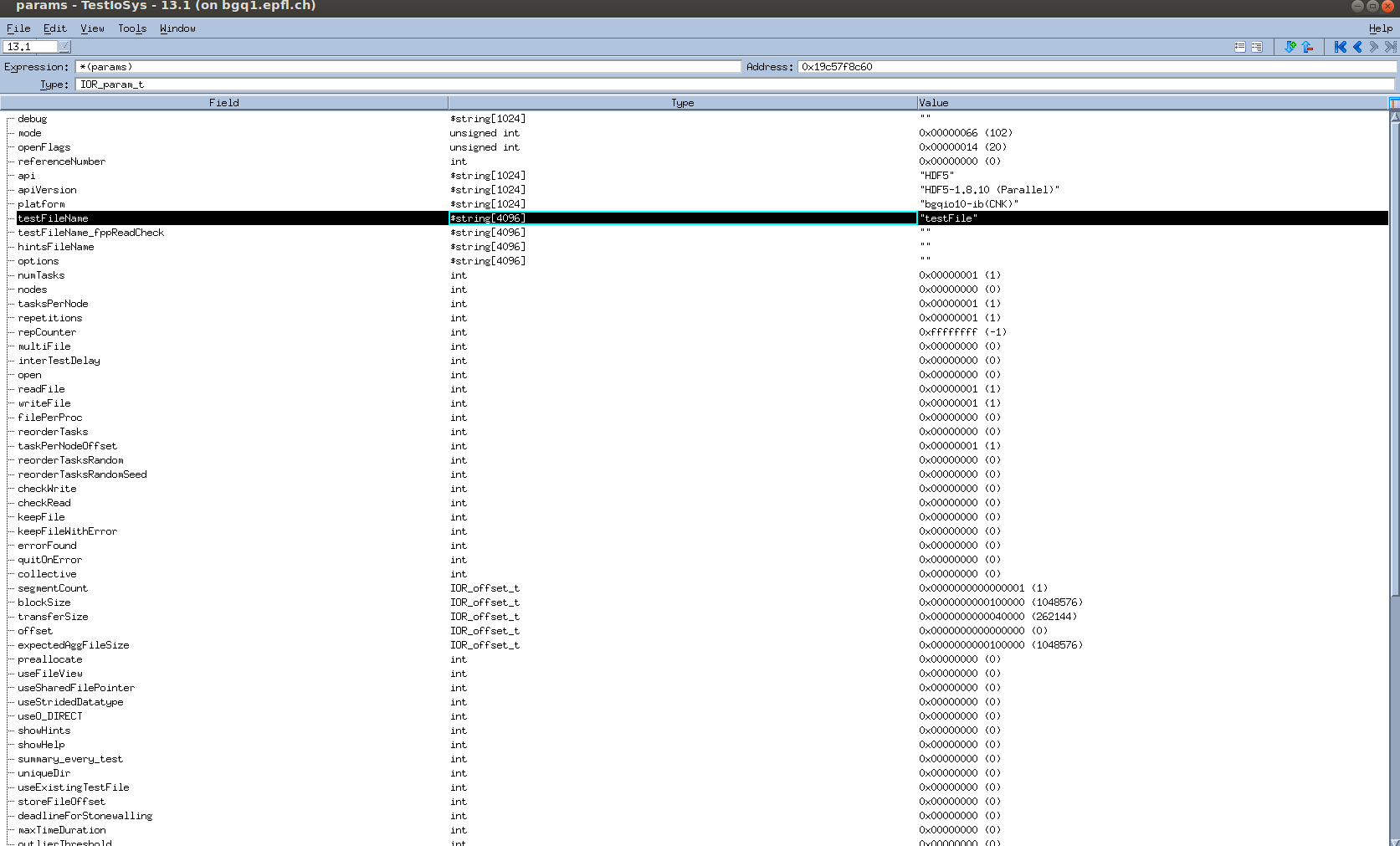
I use the latest git master branch version (v. 3.0.1)
The MPIIO and POSIX tests work fine.
Thank you for your support.
PS: As per contract with IBM, I was told by my line manager that we have to use to the version 2.10.3 of IOR... is this version fully compatible with Blue Gene Q? Any known issues?
The text was updated successfully, but these errors were encountered: