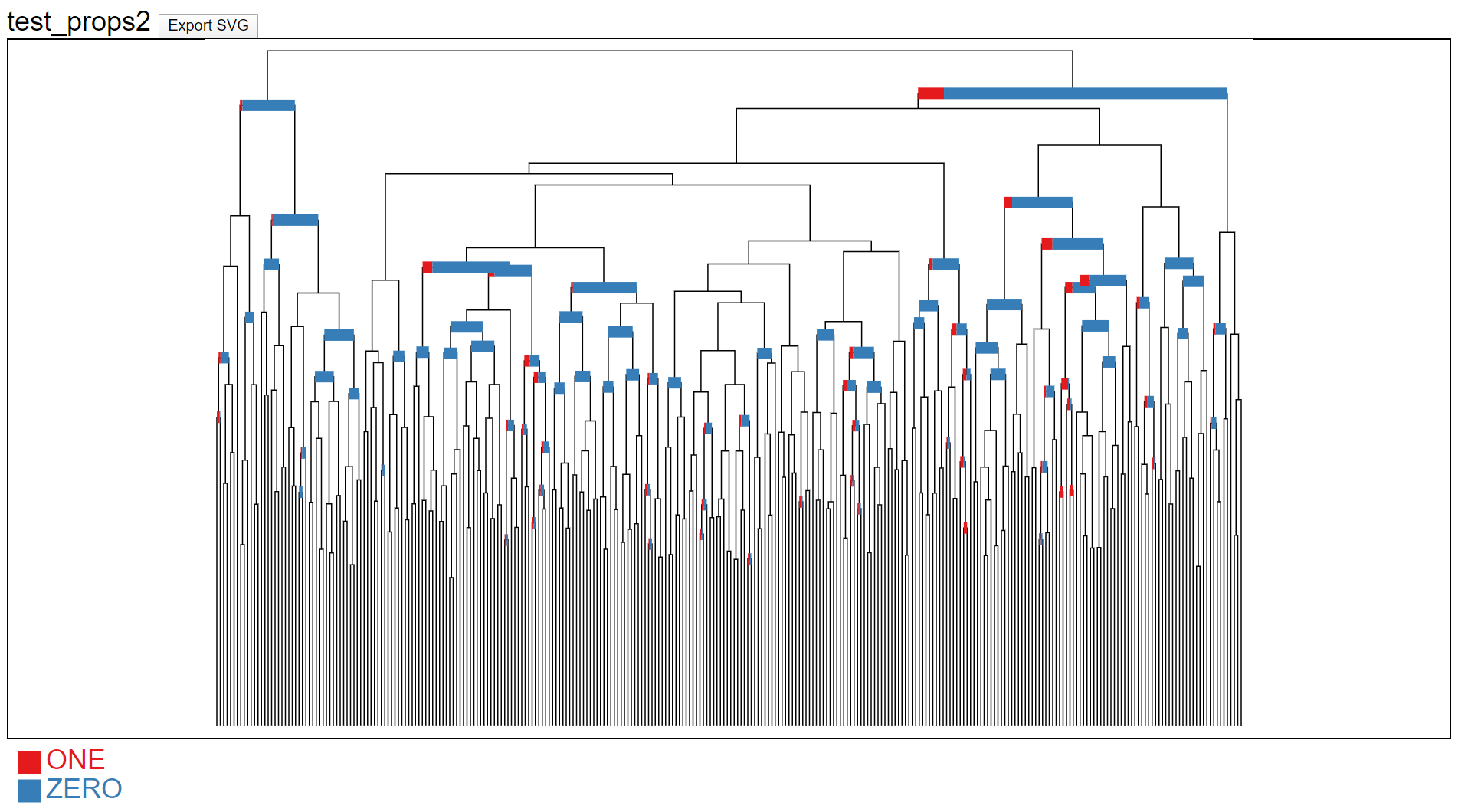
A package that is useful for clustering high-dimensional instances (e.g. T cell receptors) and testing whether clusters of instances are differentially abundant in two or more categorical conditions. The package provides d3/SVG rendering of scipy hierarchical clustering dendrograms with zooming, panning and tooltips. This uniquely allows for exploring large trees of datasets, conditioned on a categorical trait.
pip install hierdiff
import hierdiff
from scipy.spatial.distance import squareform
"""Contains categorical variable column 'trait1' and
instance counts in 'count'"""
dat, pwdist = generate_data()
res, Z = hierdiff.hcluster_tally(dat,
pwmat=squareform(pwdist),
x_cols=['trait1'],
count_col='count',
method='complete')
res = hierdiff.cluster_association_test(res, method='fishers')
"""Plot frequency of trait at nodes with p-value < 0.05"""
html = plot_hclust_props(Z, title='test_props2',
res=res, alpha=0.05, alpha_col='pvalue')