-
Notifications
You must be signed in to change notification settings - Fork 14
For Users: Web UI
Any modern web browser, such as Mozilla Firefox, Google Chrome, Safari or Microsoft Edge. Use as latest version as possible ❗
PrimerServer contains three modules that can be easily switched by the navigation tabs:

- Full (Design and Check): In this mode, it accepts target regions or target sequences (called
query) defined by users, designs primers from these regions, and searches user defined databases (calledtemplates) to defined primer specificities. - Design Only: In this mode, it just designs primers and doesn't conduct specificity check. This is usually used in NGS target sequencing panels where primer specificity is not so necessary as in legacy PCR.
- Check Only: In this mode, it accepts a set of user defined primer sequences (called
query) and check their specificity against certain databases (calledtemplates). This is usually used if users already have some primer sequences from literatures or colleagues.
Beside, there's a tab called "Visualize" where you can upload the JSON output from CLI.
By default, the module "Full (Design and Check)" would be shown in the first visit. Your browser would remember your last used module and apply it automatically in the next visit.
In the demo server, you can do a simple test using the example database to understand the usage.
In the example database, there are two sequences. They are from fragments of the D05 and A12 chromosome of the Gossypium hirsutum reference genome (NAU v1.1) with high similarity.
The menu can be selected in multiple (although in many situations single selection is enough). Be aware of the order of your selection. The first selection would be marked a star in the right column called "Selected Templates".
In the following figure, two databases were selected. In the right column called "Selected Templates", the first selected database "Example Database" was marked a star indicating it is a primary database on which specificity would be based. The other is secondary database.

Input formats are described in the CLI wiki.

- Target Region (Default): This corresponds to the
SEQUENCE_TARGETtag in Primer3. In this category, users would define a small target region (for example: SNP, INDEL or target exon/gene) and all primer pairs must flank it. Possible usage: sequencing specific sites such as INDELs, SNPs or target gene fragments. An example result: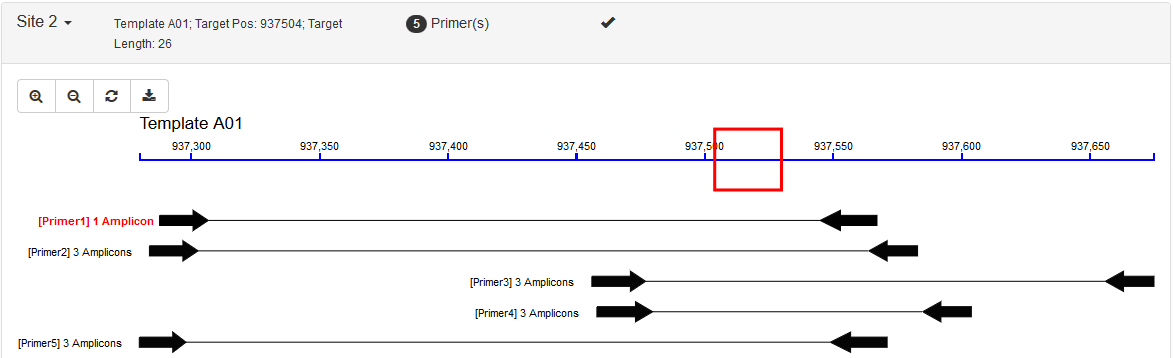
- Include Region: This corresponds to the
SEQUENCE_INCLUDED_REGIONtag in Primer3. In this category, users would define a large region (usually the whole template) and all primer pairs are picked within this region. Possible usage: quantifying specific products such as qRT-PCR. An example result: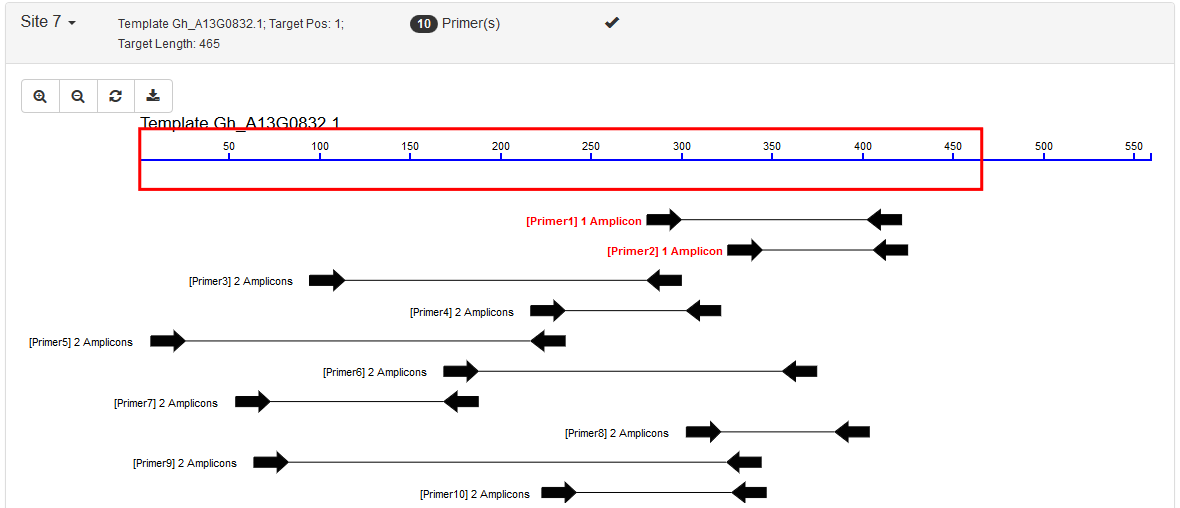
- Force 3' End: This corresponds to the
SEQUENCE_FORCE_LEFT_ENDandSEQUENCE_FORCE_RIGHT_ENDtag in Primer3. In this situation, users would define a single-base position and all left or right primers are forced to place their 3' ends to this position. Possible usage: genotyping for SNP sites. An example result: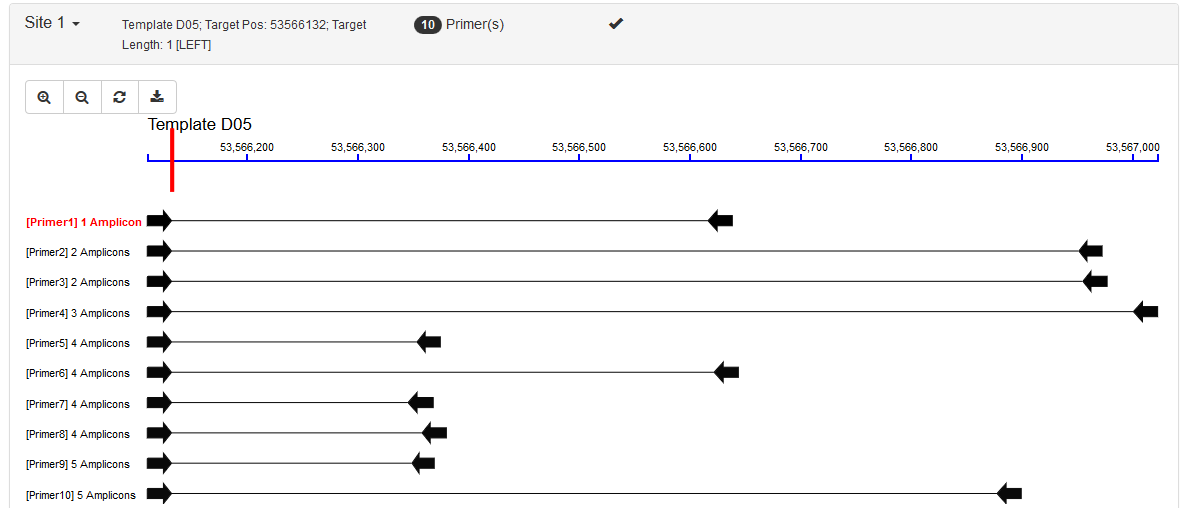
Carefully setting your product size range. They would be applied to all of the sites. If you want to give different size ranges to different sites, it is better to set them in the Input Sites.
Increase this if you want to find as many primers as possible (with longer running time), otherwise the default value is OK.
Default: 50bp~2000bp, meaning only amplicons within this range would be reported. Adjust this according to your experiment.
Default: 20°C lower. It means off-target binding primers within this range would be reported. For example, if the Tm value for a primer is 60°C, when it binds to other off-target regions, there would be lower Tm values due to mismatches and PrimerServer would report off-target bindings within Tm ranges 40°C~60°C. If no off-target bindings bindings were found in this range, this primer will be regarded as specific. Decrease this if you want to find as many primers as possible
Default: Off. If turned on, primer pairs having at least one mismatch at the 3 end position with templates would not be considered to produce off-target amplicon, even if their melting temperatures are high. Turn on this if you want to find as many primers as possible
A progress window indicating the BLAST process (the main time-consuming process) would be shown when running the app. Users can click button to stop the process if they feel it too long to finish.

Each site is listed in a collapsible panel. By default, the first panel is expanded and the others are collapsed. Users can expand the panel to see the obtained primers for each site. If a site contains unique primers, a tick is marked in the panel title.

When expanding each site, it includes two parts: an interactive graph indicating positions of the target region and its primers; and a list showing all the primers. Primers are sorted and ranked by the number of their possible PCR amplicons and then by the penalty score from Primer3.


There are two download options: Table (TSV) and JSON. Contents are described in the CLI wiki.
(1)

You can turn to the Visualize Tab, and upload the JSON file (either from previous web running or from CLI running)