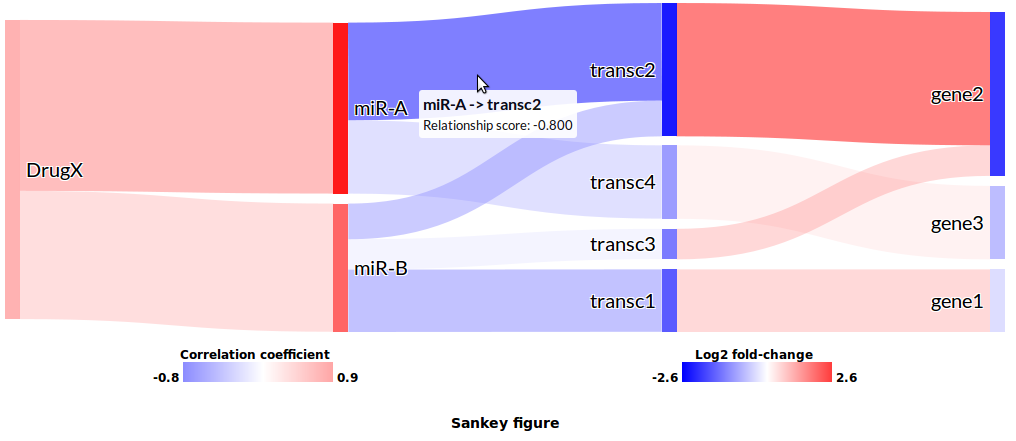
Director is an R package designed to streamline the visualization of multiple levels of interacting biological data. It utilizes the R package htmltools and customized Sankey plugin of the JavaScript library D3 (http://d3js.org) to provide an alternative to standard network visualization of biological networks. It is a fast and easy browser-enabled solution to discovering potentially interesting downstream effects or regulatory pathways. The diagrams are dynamic, interactive, and packaged as HTML files, making them highly portable and eliminating the need for third-party software. This enables a straightforward approach for scientists to interpret the data produced, and bioinformatics developers an alternative means to present relevant data.
Two types of quantitive information are all that's needed to render the diagram: values for nodes and values for paths. The package assumes nodes are molecules of interest and that the quantitative value is the feature which makes them interesting, e.g. expression fold-change, significance value, or methylation score. Similarly, paths represent a predictive or quantitative measure of the interaction between molecules (e.g. correlation, affinity score).
Quantitative values are used to render the diagram. Molecules with strong quantitative values, multiple and strong interactions are automatically emphasized in the diagram. A connected series of such molecules would, in turn, identify potentially interesting regulatory cascades -- visual information not as easily nor intuitively conveyed with standard network approaches.
Package installation instructions for R are found on http://kzouchka.github.io/Director/
Icay, K. and Liu, C. and Hautaniemi, S. (2018)
Dynamic visualization of multi-level molecular data: The Director package in R
Comput Methods Programs Biomed 153: 129--136
10.1016/j.cmpb.2017.10.013
[https://www.sciencedirect.com/science/article/pii/S0169260716306964]