-
Notifications
You must be signed in to change notification settings - Fork 1
Tutorial
In the following tutorial, we will perform an end-to-end walkthrough of a workflow that can leave you with high quality orthogroup assignments for a gene family.
A quick note up front: RD-MCL is computationally intensive – Some of the examples below could take over a week to run on a basic workstation. The good news is that all results files have been saved to the GitHub repository and links are provided in the appropriate sections, so feel free to terminate jobs after you learn how to get them running and grab the pre-calculated output to carry on with. Now without further ado, the tutorial!
- Setup
- Preamble
- Pre-generated files
- Step 1: Acquiring human protein sequences from RefSeq
- Step 2: BLAST search RefSeq for CASc domains
- Step 3: Filter BLAST hits
- Step 4: Select sequences for RD-MCL
- Step 5: Run RD-MCL
- Step 6: First look at RD-MCL results
- Step 7: Visualize RD-MCL results
- Step 8: Assessing the bird caspases
- Step 9: Assigning sequences to orthogroups
Before we get started, you will need to ensure that:
- You are working on a *NIX system (most likely Linux or Mac OS X)
- You can launch a bash shell and have at least a rudimentary understanding of how to navigate within that shell
- (if this is an issue, start by working through this excellent tutorial provided by the Software Carpentry Foundation)
- Python3 and RD-MCL are installed
- BuddySuite is installed
- The alignment program Clustal Omega is installed
- You have an active internet connection on the machine you will be running RD-MCL on
- Optional: Phylogenetic tree visualization software (FigTree is a very good choice)
All of the files downloaded/generated during this tutorial can be accessed from this list if you do not want to create them yourself.
Download files
- hsap_casp2.gb
- hsap_casp2_casc.fa
- casc_blast.csv
- casc_blast_accns.txt
- refseq_casps.gb
- arthropod_casps.fa, bird_casps.fa, fish_casps.fa, mammal_casps.fa
- rdmcl_example_output.tar.gz
The caspases will be used as our example protein family for this tutorial, because they are widely distributed across the Metazoa, there is diversity among the individual proteins, the family as a whole has already enjoyed considerable attention and annotation, and the presence of a CASc domain can be used as the minimum requirement to classify a protein as a caspase. This final point is particularly important, because it constitutes the human imposed definition of the family. Whichever family you choose to study on your own, be very clear about this definition up front!
Why? Consider that genomic plasticity is a fact of life, leading to all manner of complications when we try to reconstruct the lineage of specific genomic regions. Indels are rampant over evolutionary time, as is wholesale domain swapping, so it is rare for any two genes to be homologous along their entire length. Therefore, contemporary genetic nomenclature is largely a convenience that we use to help us understand patterns and function, not a rigorous means of documenting precise homology and descent.
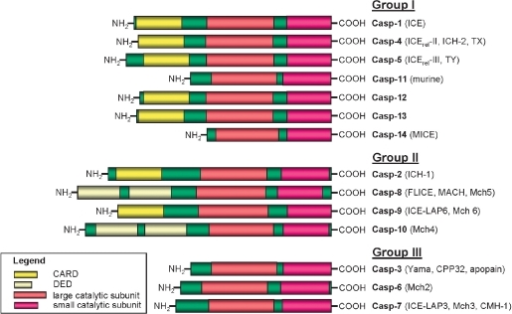
For example, consider the domain architecture of the mammalian caspases below.
Lavrik and Krammer, Acta Naturae. 2009 Apr; 1(1): 80–83
They all contain large and small catalytic domains, which presumably share homologous residues across most of their lengths, but the N-termini of each protein has some combination of DEDs and CARDs (or nothing at all). Therefore, the proteins in this 'family' are fusions of several disparate domains, and the community has decided that the catalytically active domains are the most pertinent for naming purposes. But what if the first people to study caspases were only interested in its protein binding properties? What if they decided that CARD domains were the most important feature? There are dozens of other CARD containing proteins, so one may argue that the naming convention we currently have is simply a quirk of history.
The takeaway message is to not get too hung up on a protein or gene's name, but instead to ask yourself what it is about a particular class of protein that you find the most interesting. What defining features are homologous across all of the 'family' you want to study? Once you have answered that question, you're all set to sub-classify them into useful orthogroups.
We will begin by searching the well-curated RefSeq database for the human caspase-2 sequence using DatabaseBuddy (part of BuddySuite).
Launch DatabaseBuddy and follow along. Hopefully the DatabaseBuddy commands will be intuitive, but if you're feeling lost there's a BuddySuite tutorial that you may want to go through before continuing:
$: databasebuddy
DbBuddy> database ncbi_prot
DbBuddy> search "Homo sapiens[Organism] AND caspase-2*[Protein Name] AND srcdb_refseq[PROP]"
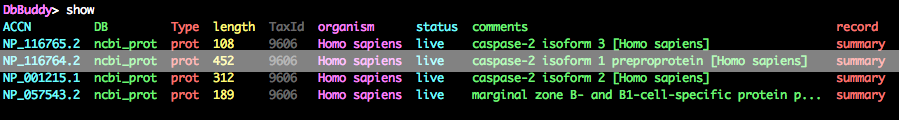
DbBuddy> show
If all has gone well, you should see a similar output as shown below after running the show command:
You'll notice that our search has pulled in a few more sequences than we wanted, but NP_116764.2 (highlighted) looks like the primary isoform, so keep that one and fetch the sequence as a GenBank file.
DbBuddy> keep "NP_116764.2"
DbBuddy> fetch
DbBuddy> format genbank
DbBuddy> write hsap_casp2.gb
DbBuddy> quit
We are starting with the human caspase-2 sequence because it is well curated. As stated in the preamble, the catalytic CASc domain is the common feature we have assigned to the family, so let's use SeqBuddy to extract that feature from caspase2 and redirect it into a FASTA file.
$: seqbuddy hsap_casp2.gb --extract_feature_sequences CASc --out_format fasta > hsap_casp2_casc.fa
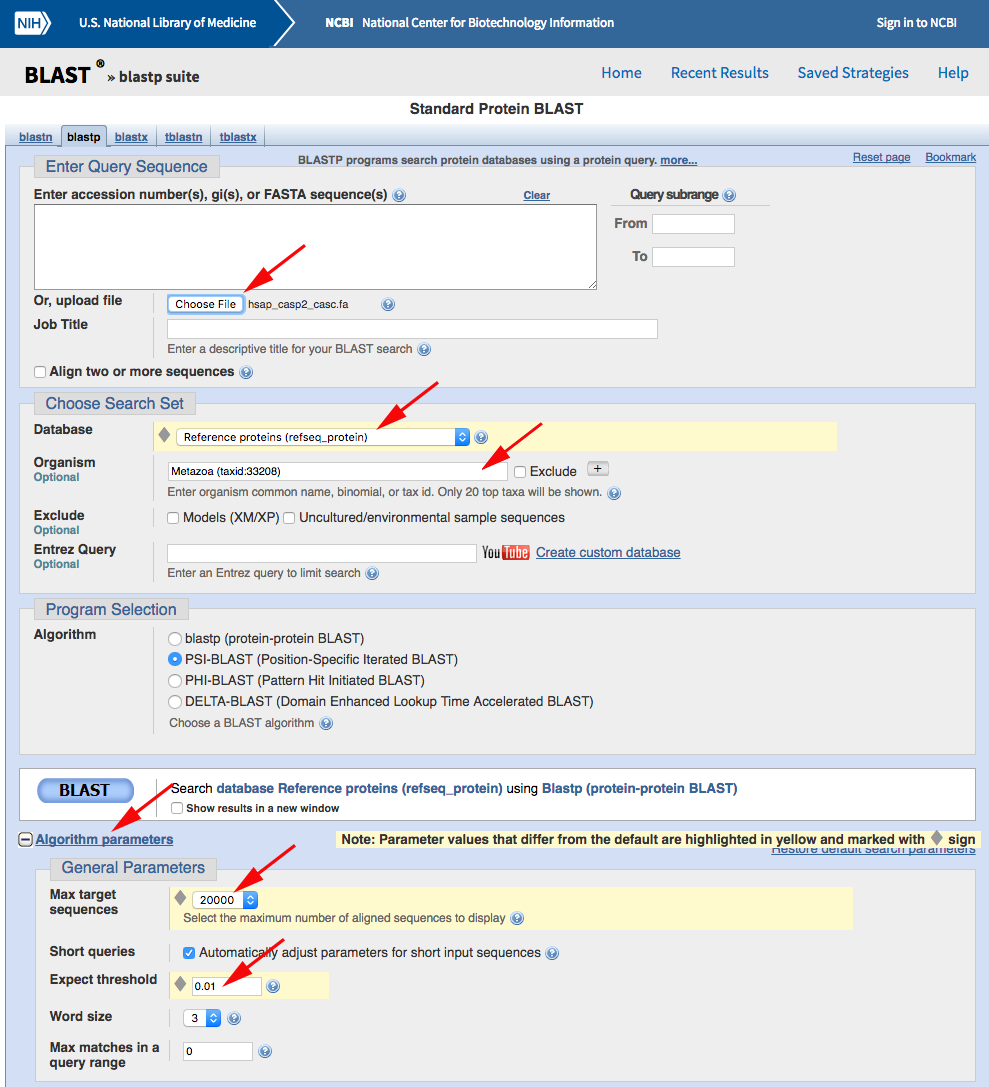
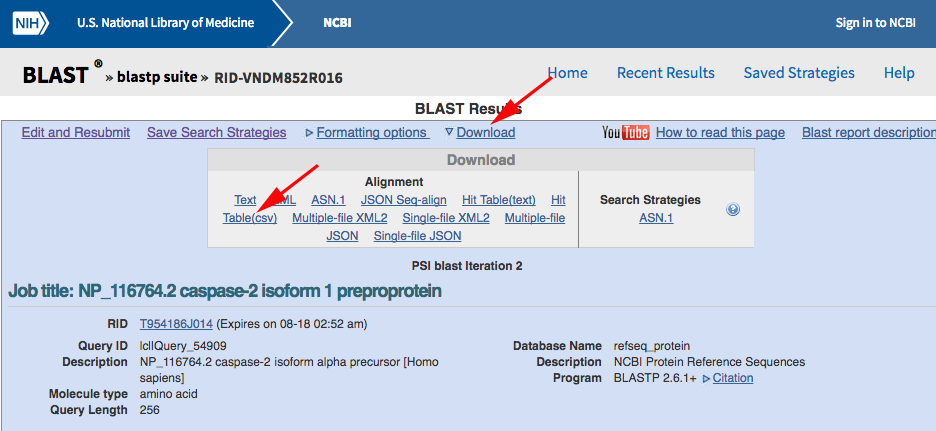
Now use PSI-BLAST to query the RefSeq database for other sequences containing a CASc domain. Navigate to the NCBI BLASTP web portal and run your search after changing the default parameters as marked with red arrows below:
You should get back almost 6000 results from the first search, but we want to iterate the PSI-BLAST a few times to pull out as many as possible. Click the "Go" button to do so:
After the third or fourth iteration, you should be seeing less than ten new records being identified, so call it quits at that point. Download the CSV hit-table:
Rename the file to something a little more meaningful.
$: mv /path/to/Downloads/<name of your file> ./casc_blast.csv
Pull out the uniq accession numbers from the hit table using the following UNIX commands and regular expression:
$: cut -f2 -d ',' casc_blast.csv | \
awk 'match($0,/[NX]P_[0-9]+(.[0-9]+)?/) {print substr($0,RSTART,RLENGTH)}' | \
sort | uniq > casc_blast_accns.txt
Now load the accessions up into DatabaseBuddy to get a better look at what was returned.
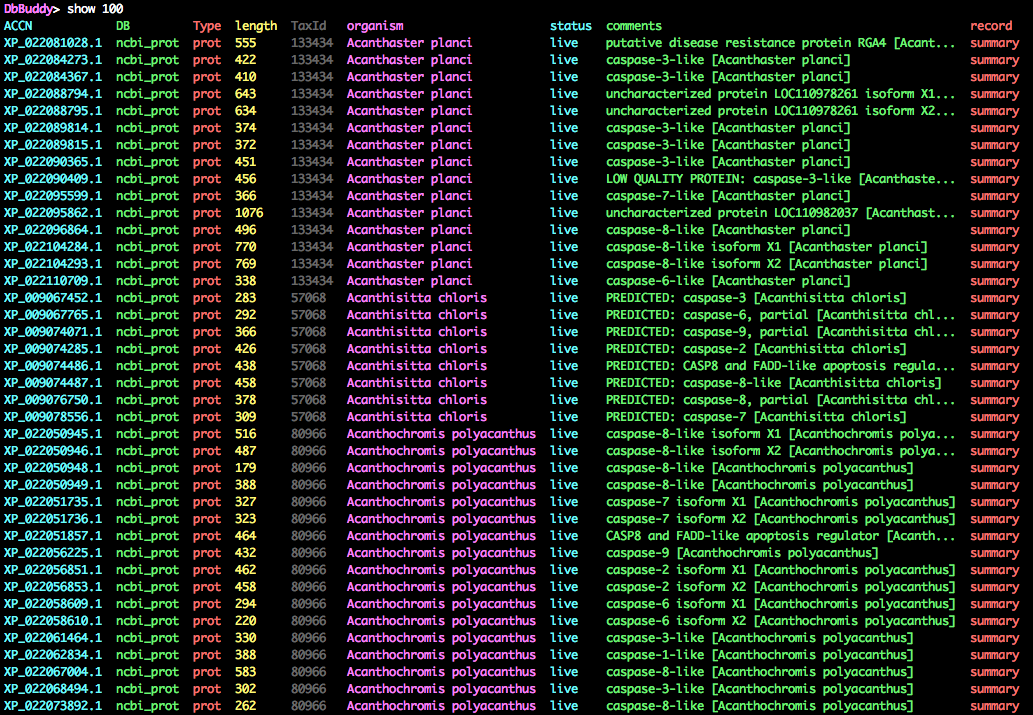
$: databasebuddy casc_blast_accns.txt -d ncbi_prot
DbBuddy> sort organism
DbBuddy> show 100
Many of the records are alternate splice variants/isoforms. Although this is useful information in many contexts, splice variants are not homologs (they are literally the same gene!), so they need to be filtered down to a representative transcript. Let's remove everything except the primary variants, and also get rid of anything marked as partial or LOW QUALITY.
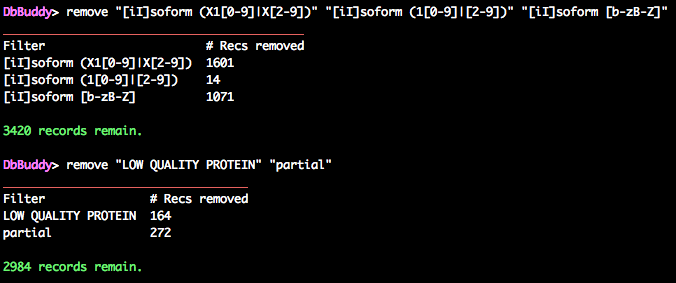
DbBuddy> remove "[iI]soform (X1[0-9]|X[2-9])" "[iI]soform (1[0-9]|[2-9])" "[iI]soform ([b-zB-Z]|A[b-zB-Z]+)"
DbBuddy> remove "LOW QUALITY PROTEIN" "partial"
This leaves just under 3,000 sequences.
If we sort the records by size, we also see a few that are very large and others that are quite small. The average caspase is around 400 residues, with a CASc domain of about 250 residues, so let's filter out the hundred-ish sequences less than 200 residues and the fifty-ish sequences that are greater than 800 residues. Large sequence can be particularly troublesome from a performance perspective, because they slow down the multiple sequence alignment considerably.
DbBuddy> sort length
DbBuddy> show 100
DbBuddy> remove (length<200)
DbBuddy> sort length reverse
DbBuddy> show 100
DbBuddy> remove (length>800)
Fetch the records and save them in GenBank format:
DbBuddy> fetch
DbBuddy> format genbank
DbBuddy> write refseq_casps.gb
DbBuddy> quit
Use the SeqBuddy command taxonomic_breakdown to visualize the taxonomic spread of the records you just curated (the integer value passed in specifies the depth of the breakdown):
$: seqbuddy refseq_casps.gb --taxonomic_breakdown 4
Eukaryota 2889
|Metazoa 2889
| |Echinodermata 34
| | |Eleutherozoa 34
| |Placozoa 1
| | |Trichoplax 1
| |Hemichordata 15
| | |Enteropneusta 15
| |Porifera 27
| | |Demospongiae 27
| |Platyhelminthes 8
| | |Trematoda 8
| |Cnidaria 80
| | |Hydrozoa 21
| | |Anthozoa 59
| |Chordata 1950
| | |Craniata 1899
| | |Tunicata 15
| | |Cephalochordata 36
| |Ecdysozoa 604
| | |Nematoda 12
| | |Scalidophora 3
| | |Arthropoda 589
| |Lophotrochozoa 170
| | |Annelida 5
| | |Brachiopoda 35
| | |Mollusca 130
The first thing that should be jumping out at you is how unbalanced the taxonomic distribution is. There is currently a strong bias in our public databases towards vertebrates and certain orders of insects, so don't be fooled into thinking there is a dearth of caspases throughout broad swaths of the Metazoa; in reality, we just haven't sequenced them yet.
RD-MCL deals reasonably well with imbalances, but the final results will be much cleaner and easier to interpret if we focus on the clades with high coverage. Let's begin by digging deeper into the distribution within the chordates:
$: seqbuddy refseq_casps.gb --keep_taxa Chordata | seqbuddy --taxonomic_breakdown 7
Eukaryota 1950
|Metazoa 1950
| |Chordata 1950
| | |Craniata 1899
| | | |Vertebrata 1899
| | | | |Chondrichthyes 11
| | | | | |Holocephali 8
| | | | | |Elasmobranchii 3
| | | | |Euteleostomi 1888
| | | | | |Actinopterygii 525
| | | | | |Amphibia 37
| | | | | |Lepidosauria 45
| | | | | |Coelacanthiformes 10
| | | | | |Archelosauria 465
| | | | | |Mammalia 806
| | |Tunicata 15
| | | |Ascidiacea 15
| | | | |Enterogona 15
| | | | | |Phlebobranchia 15
| | |Cephalochordata 36
| | | |Branchiostomidae 36
| | | | |Branchiostoma 36
| | | | | |floridae 17
| | | | | |belcheri 19
Great! It looks like three distinct groups within the Euteleostomi have high coverage: the Actinopterygii (fish), Archelosauria (turtles, crocodiles, birds), and Mammalia. Run --taxonomic_breakdown 0 on each of these clades in turn and observe the number of genes present in each species, this will give you an approximate idea as to how many orthogroups to expect:
$: seqbuddy refseq_casps.gb --keep_taxa Archelosauria | seqbuddy --taxonomic_breakdown 0
...
| | | | | | | | | | | | | | |Passeroidea 23
| | | | | | | | | | | | | | | |Estrildidae 18
| | | | | | | | | | | | | | | | |Estrildinae 18
| | | | | | | | | | | | | | | | | |Taeniopygia 10
| | | | | | | | | | | | | | | | | | |guttata 10
| | | | | | | | | | | | | | | | | |Lonchura 8
| | | | | | | | | | | | | | | | | | |striata domestica 8
| | | | | | | | | | | | | | | |Fringillidae 5
| | | | | | | | | | | | | | | | |Carduelinae 5
| | | | | | | | | | | | | | | | | |Serinus 5
| | | | | | | | | | | | | | | | | | |canaria 5
| | | | | | | | | | | | | | |Paridae 10
| | | | | | | | | | | | | | | |Pseudopodoces 6
| | | | | | | | | | | | | | | | |humilis 6
| | | | | | | | | | | | | | | |Parus 4
| | | | | | | | | | | | | | | | |major 4
| | | | | | | | | | | | | | |Pipridae 11
| | | | | | | | | | | | | | | |Manacus 6
| | | | | | | | | | | | | | | | |vitellinus 6
| | | | | | | | | | | | | | | |Lepidothrix 5
| | | | | | | | | | | | | | | | |coronata 5
| | | | | | | |Crocodylia 22
| | | | | | | | |Alligatoridae 13
| | | | | | | | | |Alligatorinae 13
| | | | | | | | | | |Alligator 13
| | | | | | | | | | | |sinensis 8
| | | | | | | | | | | |mississippiensis 5
| | | | | | | | |Longirostres 9
| | | | | | | | | |Gavialidae 4
...
By my guess, we should anticipate around 10 major orthogroups for each of these clades. If you observe 20+ genes in each species for your pet protein family, than you would expect closer to twenty, and likewise, expect much fewer if you only see two or three genes per species. It's just a rough estimate.
Let's prepare each group for RD-MCL:
$: seqbuddy refseq_casps.gb --keep_taxa Actinopterygii | seqbuddy --prepend_organism --out_format fasta > fish_casps.fa
$: seqbuddy refseq_casps.gb --keep_taxa Archelosauria | seqbuddy --prepend_organism --out_format fasta > bird_casps.fa
$: seqbuddy refseq_casps.gb --keep_taxa Mammalia | seqbuddy --prepend_organism --out_format fasta > mammal_casps.fa
Similarly, have a look at the Arthropoda:
$: seqbuddy refseq_casps.gb --keep_taxa Arthropoda | seqbuddy --taxonomic_breakdown 6
Total: 599
Eukaryota 589
|Metazoa 589
| |Ecdysozoa 589
| | |Arthropoda 589
| | | |Crustacea 12
| | | | |Malacostraca 12
| | | |Hexapoda 553
| | | | |Insecta 544
| | | | |Collembola 9
| | | |Chelicerata 24
| | | | |Merostomata 7
| | | | |Arachnida 17
Insects are making up the vast majority, so zoom in on them:
seqbuddy refseq_casps.gb --keep_taxa Insecta | seqbuddy --taxonomic_breakdown 10
Total: 554
Eukaryota 544
|Metazoa 544
| |Ecdysozoa 544
| | |Arthropoda 544
| | | |Hexapoda 544
| | | | |Insecta 544
| | | | | |Pterygota 544
| | | | | | |Neoptera 544
| | | | | | | |Polyneoptera 3
| | | | | | | | |Dictyoptera 3
| | | | | | | |Paraneoptera 25
| | | | | | | | |Psocodea 4
| | | | | | | | |Hemiptera 21
| | | | | | | |Holometabola 516
| | | | | | | | |Lepidoptera 45
| | | | | | | | |Coleoptera 42
| | | | | | | | |Diptera 282
| | | | | | | | |Hymenoptera 147
Most of the coverage is in the Diptera (flies) and Hymenoptera (wasps/bees/ants), so grab out those two clades for further analysis as well:
$: seqbuddy refseq_casps.gb --keep_taxa Diptera | seqbuddy --prepend_organism --out_format fasta > diptera_casps.fa
$: seqbuddy refseq_casps.gb --keep_taxa Hymenoptera | seqbuddy --prepend_organism --out_format fasta > hymenoptera_casps.fa
Note that in each of the commands above, there is a final call to the SeqBuddy function prepend_organism. This creates and appends a species identifier (first character of the genus name, first three characters from the species, and an integer if there would otherwise be a naming conflict) to each gene in the data-set, as is required by RD-MCL.
RD-MCL builds all-by-all similarity graphs among the sequences it is provided; this step can be very computationally expensive because these types of problems grow quadratically (i.e., doubling the number of sequences quadruples the runtime, at minimum). On my workstation, which runs 12 hyper-threaded processors, I can process 100 sequences in a few hours, while 800 sequences takes more than a week. If you have access to a cluster, I strongly recommend parallelizing the job for more than a couple hundred sequences.
Running RD-MCL serially is straightforward; at minimum, just call the program and tell it where the sequence file is:
$: rdmcl bird_casps.fa
RD-MCL has a couple dozen optional parameters you can set if you want to modify the run. For a list of those options call rdmcl --help or see the dedicated wiki page. We'll use some of these parameters to add more informative names to our output directory and final clusters.
$: rdmcl bird_casps.fa --outdir bird --group_name bird
$: rdmcl mammal_casps.fa --outdir mammal --group_name mamm
$: rdmcl fish_casps.fa --outdir fish --group_name fish
$: rdmcl diptera_casps.fa --outdir diptera --group_name dipt
$: rdmcl hymenoptera_casps.fa --outdir hymenoptera --group_name hyme
If you are following along with your own data, it will probably take a while for the job(s) to finish. If you are following along with the caspase example, you can grab the pre-computed results from here (they will be used in the following steps).
After a run has complete, the output directory should have a collection of files and folders that looks like this:
$: ls bird/
alignments/ hmm/ mcmcmc/
psi_pred/ placement.log rdmcl.log
final_clusters.txt paralog_cliques sim_scores/
sqlite_db.sqlite
| File/folder | Description |
|---|---|
| alignments/ | The MSAs used to generate the all-by-all similarity scores for each final cluster |
| mcmcmc/ | The output of each mcmcmc chain for each final cluster |
| psi_pred/ | Secondary structure .ss2 files for each input sequence |
| hmm/ | Collection of hidden Markov model files |
| placement.log | Final sequence placement into clusters log |
| rdmcl.log | Description of the run |
| final_clusters.txt | Your main result! |
| paralog_cliques | List of RBH paralogs extracted from the dataset before running RDMCL |
| sim_scores/ | All-by-all similarity graphs for each final cluster |
| sqlite_db.sqlite | SQLlight database with all of the MSAs and similarity graphs from the run |
The two files you will be most interested in, at least at first, are rdmcl.log and final_clusters.txt
The rdmcl.log file gives a full breakdown of all parameters used to generate your RD-MCL run. This will allow you to keep track of what you did and reproduce the result if necessary. Each part of this file is explained in greater detail in a separate wiki page.
$: cat bird/rdmcl.log
*************************** Recursive Dynamic Markov Clustering ****************************
RD-MCL version 1.0.4
Public Domain Notice
--------------------
This is free software; see the LICENSE for further details.
There is NO warranty; not even for MERCHANTABILITY or FITNESS FOR A
PARTICULAR PURPOSE.
Questions/comments/concerns can be directed to Steve Bond, steve.bond@nih.gov
--------------------
********************************************************************************************
Function call: /home/bondsr/bioinf_tools/rdmcl bird_casps.fa -o bird_casps -rs 1 -q -cpu 16 -wdb /data/bondsr/workers/ -psi psipred
Date: 11:35-21-12-2017
** Environment setup **
Random seed used for this run: 1
Working directory: /data/bondsr/caspases
Output directory:
/data/bondsr/caspases/bird_casps
SeqBuddy version: 1.3.0
AlignBuddy version: 1.3.0
Alignment method: CLUSTALO 1.2
Alignment calls: $: clustalo --pileup
TrimAl values: ['gappyout', 0.5, 0.75, 0.9, 0.95, 'clean']
Launching SQLite Daemons
Checking that the format of all sequence ids matches 'taxa-gene'
465 sequences PASSED
68 unique taxa present
** PSI-Pred **
RESUME: All PSI-Pred .ss2 files found
** All-by-all graph **
gap open penalty: -5
gap extend penalty: 0
Generating initial all-by-all similarity graph (107880 comparisons)
written to: bird_casps/sim_scores/complete_all_by_all.scores
-- finished in 9 min, 5 sec --
Base cluster score: 236.2077
** Recursive MCL **
Auto detect MCMCMC convergence
Gelman-Rubin convergence breakpoint: 1.1
Number of MCMC chains: 3
Number of Metropolis-Hastings walkers per chain: 3
Total MCL runs: 4503
-- finished in 9 hrs, 36 min, 36 sec --
** HMM-based sequence-to-cluster reassignment **
-- finished in 11 min, 58 sec --
Placing any collapsed paralogs into their respective clusters
Preparing final_clusters.txt
Total execution time: 9 hrs, 57 min, 39 sec
Final score: 1068.3789
Clusters written to: bird_casps/final_clusters.txt
Probably even more interesting to you, are the results in final_clusters.txt:
The first column is the group name assigned to the cluster. The second column is the final score of the cluster, based on a diminishing returns algorithm. The remaining tab separated columns are the sequence names for the members of that cluster.
$: cat bird/final_clusters.txt
bird_0_0_0 161.6026 Acar-XP_010171751.1 Achl-XP_009067452.1 Achr-XP_011577228.1 Acyg-XP_013035279.1 Afor-XP_009275633.1 Amis-XP_006263939.1 Apla-XP_005030551.1 Asin-XP_006026683.1 Breg-XP_010308818.1 Brhi-XP_010129400.1 Cann-XP_008490097.1 Cbra-XP_008639693.1 Ccan-XP_009563972.1 Ccri-XP_009697353.1 Cjap-XP_015716892.1 Cliv-XP_005500235.1 Cmyd-XP_007054526.1 Cpel-XP_010000752.1 Cpor-XP_019390228.1 Cpug-XP_014800971.1 Cvoc-XP_009890080.1 Egar-XP_009640697.1 Falb-XP_005045177.1 Fche-XP_005445415.1 Fgla-XP_009570651.1 Fper-XP_005230888.1 Gfor-XP_005415590.1 Ggal-NP_990056.1 Ggan-XP_019377451.1 Gste-XP_009810205.1 Halb-XP_009922022.1 Lcor-XP_017664322.1 Ldis-XP_009958677.1 Lstr-XP_021399050.1 Mnub-XP_008944070.1 Mund-XP_005149203.1 Muni-XP_010182555.1 Mvit-XP_008923018.1 Nmel-XP_021249038.1 Nnip-XP_009473093.1 Nnot-XP_010009471.1 Ohoa-XP_009932264.1 Pade-XP_009328950.1 Pcar-XP_009512955.1 Pcri-XP_009479867.1 Pgut-XP_010078873.1 Phum-XP_005517714.1 Plep-XP_010281454.1 Pmaj-XP_015479949.1 Ppub-XP_009909179.1 Svul-XP_014745174.1 Talb-XP_009970400.1 Tery-XP_009988707.1 Tgut1-XP_002191294.1 Tgut1-XP_012428936.1 Zalb-XP_005483896.1
bird_0_1 115.6107 Acar-XP_010163578.1 Acyg-XP_013040763.1 Afor-XP_019326128.1 Apla-XP_021128354.1 Asin-XP_006025610.2 Avit-XP_009875221.1 Breg-XP_010310914.1 Brhi-XP_010134615.1 Cann-XP_008497831.1 Ccan-XP_009563276.1 Ccor-XP_010410023.1 Ccri-XP_009699574.1 Cjap-XP_015717471.1 Cmac-XP_010124189.1 Cmyd-XP_007054543.1 Cpel-XP_010002388.1 Cpic-XP_005287972.1 Cstr-XP_010200177.1 Cvoc-XP_009883787.1 Egar-XP_009647487.1 Fgla-XP_009578173.1 Fper-XP_013155138.1 Ggal-NP_990057.1 Halb-XP_009924247.1 Hleu-XP_010584548.1 Lcor-XP_017661517.1 Ldis-XP_009947440.1 Mgal-XP_010708306.2 Mund-XP_005148683.1 Muni-XP_010187950.1 Mvit-XP_017927808.1 Nmel-XP_021252130.1 Nnip-XP_009472930.1 Nnot-XP_010015536.1 Ohoa-XP_009930434.1 Pade-XP_009325341.1 Pcar-XP_009511931.1 Pcri-XP_009483599.1 Pgut-XP_010082525.1 Phum-XP_005517572.1 Plep-XP_010290767.1 Pmaj-XP_015480600.1 Ppub-XP_009903163.1 Psin-XP_014427079.1 Talb-XP_009966306.1 Tery-XP_009977213.1 Tgut1-XP_002197219.1
bird_0_2 114.1264 Aaus-XP_013807673.1 Acar-XP_010167151.1 Achl-XP_009074486.1 Achr-XP_011574988.1 Avit-XP_009876193.1 Brhi-XP_010144488.1 Cann-XP_008494158.1 Ccan-XP_009558080.1 Ccor-XP_010404899.1 Ccri-XP_009707893.1 Cjap-XP_015723629.1 Cmac-XP_010123198.1 Cpel-XP_010005644.1 Cpic-XP_005314340.1 Cpug-XP_014809790.1 Cstr-XP_010196023.1 Cvoc-XP_009888482.1 Egar-XP_009641851.1 Ehel-XP_010145466.1 Falb-XP_005049244.1 Fgla-XP_009573771.1 Gfor-XP_005415995.1 Ggal-NP_001189391.1 Gste-XP_009807246.1 Hleu-XP_010563070.1 Ldis-XP_009950008.1 Lstr-XP_021411077.1 Mgal-XP_010711730.1 Mund-XP_005145602.2 Muni-XP_010189510.1 Nnip-XP_009460549.1 Nnot-XP_010009278.1 Ohoa-XP_009938107.1 Pcar-XP_009506123.1 Pcri-XP_009479033.1 Pgut-XP_010076877.1 Phum-XP_005519962.2 Plep-XP_010285479.1 Ppub-XP_009894784.1 Psin-XP_006118011.1 Scan-XP_009086337.1 Svul-XP_014734929.1 Talb-XP_009974979.1 Tery-XP_009976784.1 Tgut1-XP_002188318.3 Zalb-XP_005484242.1
bird_0_3 111.1932 Aaus-XP_013797196.1 Achl-XP_009078556.1 Acyg-XP_013030032.1 Apla-XP_005014501.1 Avit-XP_009876249.1 Breg-XP_010295946.1 Cann-XP_008488344.1 Cbra-XP_008632648.1 Ccan-XP_009568871.1 Ccri-XP_009697487.1 Cjap-XP_015722817.1 Cmac-XP_010117445.1 Cpic-XP_005293940.1 Cpug-XP_014814053.1 Cstr-XP_010198191.1 Cvoc-XP_009879870.1 Egar-XP_009634188.1 Ehel-XP_010155261.1 Fgla-XP_009578529.1 Gfor-XP_005416425.1 Ggal-XP_421764.3 Gste-XP_009805143.1 Halb-XP_009923386.1 Ldis-XP_009948704.1 Lstr-XP_021381698.1 Mgal-XP_019473523.1 Mvit-XP_017924402.1 Nmel-XP_021255526.1 Nnip-XP_009474328.1 Nnot-XP_010010881.1 Ohoa-XP_009932919.1 Pade-XP_009324216.1 Pcar-XP_009508983.1 Pcri-XP_009483464.1 Phum-XP_005520811.1 Plep-XP_010288794.1 Ppub-XP_009906461.1 Psin-XP_006135034.1 Scam-XP_009666939.1 Scan-XP_009085424.1 Svul-XP_014728008.1 Talb-XP_009965879.1 Tgut1-XP_012430035.1 Tgut2-XP_010215551.1 Zalb-XP_005481147.1
bird_0_4 103.7729 Aaus-XP_013807671.1 Acar-XP_010165007.1 Achr-XP_011574945.1 Afor-XP_009282933.1 Apla-XP_012950189.1 Avit-XP_009876852.1 Breg-XP_010296000.1 Cann-XP_008494202.1 Ccan-XP_009558108.1 Ccri-XP_009701338.1 Cmac-XP_010125639.1 Cmyd-XP_007072004.1 Cpel-XP_010005642.1 Cpug-XP_014810034.1 Cvoc-XP_009888483.1 Egar-XP_009641850.1 Ehel-XP_010147131.1 Falb-XP_005049234.1 Fgla-XP_009583496.1 Gfor-XP_005415994.1 Ggal-XP_421936.4 Gste-XP_009807895.1 Halb-XP_009924272.1 Ldis-XP_009949374.1 Lstr-XP_021411054.1 Mgal-XP_010711743.1 Mnub-XP_008941675.1 Mund-XP_005140102.1 Mund-XP_005144727.1 Nnip-XP_009460550.1 Nnot-XP_010017544.1 Ohoa-XP_009938108.1 Pade-XP_009330078.1 Pcar-XP_009500813.1 Pcri-XP_009483191.1 Phum-XP_005519961.1 Plep-XP_010288754.1 Pmaj-XP_015489704.1 Ppub-XP_009894800.1 Scam-XP_009682549.1 Scan-XP_009086338.1 Talb-XP_009963924.1 Tgut1-XP_002188263.2 Tgut2-XP_010222872.1 Zalb-XP_005484241.1
bird_0_5 96.5389 Achl-XP_009074285.1 Afor-XP_009285422.1 Avit-XP_009872031.1 Breg-XP_010308307.1 Brhi-XP_010140546.1 Cann-XP_008497332.1 Ccan-XP_009564969.1 Ccor-XP_010394452.1 Ccri-XP_009706668.1 Cmyd-XP_007066518.1 Cpel-XP_009996635.1 Cpic-XP_005281708.1 Cvoc-XP_009891089.1 Egar-XP_009637435.1 Falb-XP_005038461.2 Fche-XP_014143045.1 Fper-XP_013153061.1 Gfor-XP_005430019.1 Ggal-NP_001161173.1 Gste-XP_009806633.1 Halb-XP_009910597.1 Hleu-XP_010565918.1 Ldis-XP_009946982.1 Lstr-XP_021404675.1 Mgal-XP_010715921.1 Mund-XP_005144852.1 Muni-XP_010184195.1 Nmel-XP_021251148.1 Nnip-XP_009459864.1 Nnot-XP_010011239.1 Ohoa-XP_009930304.1 Pade-XP_009326897.1 Phum-XP_005523866.1 Plep-XP_010281146.1 Pmaj-XP_015496309.1 Ppub-XP_009897931.1 Scam-XP_009683457.1 Scan-XP_009100903.1 Svul-XP_014747857.1 Talb-XP_009968311.1 Tgut1-XP_002192067.2 Zalb-XP_005495024.1
bird_0_6 95.903 Aaus-XP_013807675.1 Acar-XP_010165008.1 Achl-XP_009074487.1 Afor-XP_009282932.1 Apla-XP_021124568.1 Avit-XP_009876853.1 Breg-XP_010296001.1 Cann-XP_008494157.1 Ccan-XP_009558078.1 Ccor-XP_010404904.1 Ccri-XP_009701337.1 Cjap-XP_015723635.1 Cliv-XP_021155180.1 Cmac-XP_010125640.1 Cpel-XP_010005641.1 Cpug-XP_014818396.1 Cstr-XP_010197451.1 Ehel-XP_010147130.1 Falb-XP_005049235.1 Fgla-XP_009583493.1 Gfor-XP_005415993.1 Ggal-NP_001038154.1 Gste-XP_009807896.1 Halb-XP_009924273.1 Lcor-XP_017693623.1 Ldis-XP_009949365.1 Lstr-XP_021411019.1 Mgal-XP_010711755.1 Muni-XP_010177246.1 Mvit-XP_008929353.1 Nmel-XP_021256512.1 Nnot-XP_010017555.1 Pade-XP_009331031.1 Pcar-XP_009500812.1 Pcri-XP_009482626.1 Pgut-XP_010071041.1 Plep-XP_010288743.1 Ppub-XP_009894798.1 Scam-XP_009682550.1 Svul-XP_014734940.1 Talb-XP_009963925.1 Tgut1-XP_012430417.1
bird_0_7 86.1436 Aaus-XP_013807775.1 Achr-XP_011575010.1 Acyg-XP_013043041.1 Afor-XP_009282931.1 Asin-XP_006022169.1 Brhi-XP_010133584.1 Cann-XP_008494156.1 Cjap-XP_015723633.1 Cliv-XP_021155179.1 Cmyd-XP_007059506.1 Cpel-XP_010005640.1 Cpug-XP_014809874.1 Cvoc-XP_009888484.1 Egar-XP_009641849.1 Falb-XP_005049226.1 Fche-XP_014133274.1 Fper-XP_013161015.1 Gfor-XP_005415992.1 Ggal-NP_989923.1 Halb-XP_009919268.1 Hleu-XP_010563140.1 Lcor-XP_017689443.1 Lstr-XP_021411142.1 Mgal-XP_003207543.1 Muni-XP_010177244.1 Mvit-XP_017927167.1 Nmel-XP_021253377.1 Nnip-XP_009460551.1 Ohoa-XP_009938109.1 Pade-XP_009330066.1 Ppub-XP_009894783.1 Scam-XP_009682548.1 Scan-XP_009086335.1 Svul-XP_014734964.1 Talb-XP_009961244.1 Tgut1-XP_002190143.1 Tgut2-XP_010222877.1 Zalb-XP_005484240.1
bird_0_8 62.6434 Aaus-XP_013808908.1 Achr-XP_011583470.1 Afor-XP_009288035.1 Apla-XP_021124881.1 Asin-XP_006037744.1 Cann-XP_008489460.1 Ccan-XP_009560717.1 Cjap-XP_015737946.1 Cmyd-XP_007067800.1 Cpel-XP_010003525.1 Cpor-XP_019397069.1 Cvoc-XP_009888283.1 Egar-XP_009644820.1 Falb-XP_005057807.1 Fper-XP_013155413.1 Gfor-XP_014165459.1 Ggal-XP_424580.5 Ggan-XP_019367434.1 Hleu-XP_010575587.1 Lcor-XP_017687439.1 Lstr-XP_021389475.1 Mgal-XP_010721334.2 Mund-XP_005145535.1 Mvit-XP_017934813.1 Nmel-XP_021272957.1 Ohoa-XP_009940847.1 Pcri-XP_009484245.1 Ppub-XP_009905124.1 Psin-XP_006132789.1 Scam-XP_009687912.1 Tgut1-XP_002190691.2 Tgut2-XP_010226293.1 Zalb-XP_014128778.1
bird_0_9 51.3829 Aaus-XP_013798363.1 Achr-XP_011592833.1 Afor-XP_009274713.1 Breg-XP_010307140.1 Ccan-XP_009560405.1 Ccri-XP_009703294.1 Cliv-XP_005515064.1 Cmac-XP_010125475.1 Cpug-XP_014808566.1 Cstr-XP_010193785.1 Cvoc-XP_009887176.1 Egar-XP_009643261.1 Fgla-XP_009573990.1 Fper-XP_005236179.1 Hleu-XP_010584226.1 Mnub-XP_008937577.1 Mund-XP_005139687.1 Muni-XP_010189744.1 Nmel-XP_021270990.1 Nnip-XP_009463110.1 Nnot-XP_010022076.1 Pade-XP_009321893.1 Pcar-XP_009511202.1 Pgut-XP_010085267.1 Plep-XP_010282153.1 Scam-XP_009676466.1 Talb-XP_009970325.1
bird_0_10 30.2185 Achr-XP_011573657.1 Acyg-XP_013056451.1 Afor-XP_009283935.1 Breg-XP_010306058.1 Ccan-XP_009567978.1 Cpel-XP_010000563.1 Cpug-XP_014814213.1 Cstr-XP_010197422.1 Cvoc-XP_009881756.1 Egar-XP_009632096.1 Fgla-XP_009572183.1 Fper-XP_005235802.1 Gste-XP_009807903.1 Halb-XP_009921492.1 Mgal-XP_003214157.1 Nmel-XP_021230580.1 Nnip-XP_009474633.1 Pade-XP_009333346.1 Talb-XP_009966940.1
bird_0_11 12.4519 Amis-XP_006268804.1 Asin-XP_006031223.1 Cmyd-XP_007067525.1 Cpic-XP_005313463.1 Cpor-XP_019412216.1 Ggan-XP_019356914.1 Psin-XP_006129195.1
bird_0_12 8.2432 Amis-XP_006267994.1 Asin-XP_006033845.1 Cmyd-XP_007054624.1 Cpic-XP_008173016.1 Cpor-XP_019402013.1 Psin-XP_014427356.1
bird_0_14 8.2059 Amis-XP_006263127.1 Asin-XP_006037942.1 Cpor-XP_019393144.1 Ggan-XP_019364464.1
bird_0_13_0 3.6958 Cmyd-XP_007070748.1 Cpic-XP_005278627.1 Psin-XP_006121821.1
bird_0_15 2.1732 Cmyd-XP_007072003.1 Psin-XP_014434339.1
bird_0_16 3.3456 Amis-XP_006272609.1 Asin-XP_006022170.1
bird_0_13_1 1.1275 Psin-XP_006121825.1
Use the group_by_cluster program (bundled with RD-MCL) to map the definition line from the original sequence file back onto the gene ids and print them out as a list:
$: group_by_cluster bird
bird_0_0_0
Acar-XP_010171751.1 PREDICTED: caspase-3 [Antrostomus carolinensis]
Achl-XP_009067452.1 PREDICTED: caspase-3 [Acanthisitta chloris]
Achr-XP_011577228.1 PREDICTED: caspase-3 [Aquila chrysaetos canadensis]
Acyg-XP_013035279.1 PREDICTED: caspase-3 [Anser cygnoides domesticus]
Afor-XP_009275633.1 PREDICTED: caspase-3 [Aptenodytes forsteri]
Amis-XP_006263939.1 PREDICTED: caspase-3 [Alligator mississippiensis]
......
bird_0_13_0
Cmyd-XP_007070748.1 PREDICTED: caspase-14-like [Chelonia mydas]
Cpic-XP_005278627.1 PREDICTED: caspase-14-like [Chrysemys picta bellii]
Psin-XP_006121821.1 PREDICTED: caspase-14-like [Pelodiscus sinensis]
bird_0_15
Cmyd-XP_007072003.1 PREDICTED: caspase-8-like [Chelonia mydas]
Psin-XP_014434339.1 PREDICTED: caspase-8-like [Pelodiscus sinensis]
bird_0_16
Amis-XP_006272609.1 PREDICTED: caspase-8 [Alligator mississippiensis]
Asin-XP_006022170.1 PREDICTED: caspase-8-like [Alligator sinensis]
bird_0_13_1
Psin-XP_006121825.1 PREDICTED: caspase-14-like [Pelodiscus sinensis]
This is an excellent way to get an initial feel for how clean the orthogroups are, how well these genes have been annotated in RefSeq, and whether there are any previously unrecognized orthogroups. For example, almost all of the bird caspase records are annotated and almost all of the annotations are sorted perfectly into the clusters returned by RD-MCL. Out of the 465 sequences analyzed, one is labelled as uncharacterized and four are labelled differently from the majority of sequences in their cluster. All of these are listed below:
bird_0_0_0 (putative caspase-3)
.....
Tery-XP_009988707.1 PREDICTED: caspase-3 [Tauraco erythrolophus]
Tgut1-XP_002191294.1 PREDICTED: caspase-3-like [Taeniopygia guttata]
Tgut1-XP_012428936.1 PREDICTED: uncharacterized protein LOC100222232 [Taeniopygia guttata]
Zalb-XP_005483896.1 PREDICTED: caspase-3 [Zonotrichia albicollis]
bird_0_7 (putative caspase-8)
.....
Falb-XP_005049226.1 PREDICTED: caspase-8-like [Ficedula albicollis]
Fche-XP_014133274.1 PREDICTED: caspase-8-like [Falco cherrug]
Fper-XP_013161015.1 PREDICTED: CASP8 and FADD-like apoptosis regulator [Falco peregrinus]
Gfor-XP_005415992.1 PREDICTED: caspase-8-like [Geospiza fortis]
Ggal-NP_989923.1 caspase-8 [Gallus gallus]
.....
bird_0_10 (putative caspase-3)
Achr-XP_011573657.1 PREDICTED: caspase-3-like [Aquila chrysaetos canadensis]
Acyg-XP_013056451.1 PREDICTED: caspase-10-like [Anser cygnoides domesticus]
Afor-XP_009283935.1 PREDICTED: caspase-3 [Aptenodytes forsteri]
Breg-XP_010306058.1 PREDICTED: caspase-3-like [Balearica regulorum gibbericeps]
.....
bird_0_11 (caspase-3/7?)
Amis-XP_006268804.1 PREDICTED: caspase-3 [Alligator mississippiensis]
Asin-XP_006031223.1 PREDICTED: caspase-3-like [Alligator sinensis]
Cmyd-XP_007067525.1 PREDICTED: caspase-3-like [Chelonia mydas]
Cpic-XP_005313463.1 PREDICTED: caspase-7-like [Chrysemys picta bellii]
Cpor-XP_019412216.1 PREDICTED: caspase-3-like [Crocodylus porosus]
Ggan-XP_019356914.1 PREDICTED: caspase-3-like [Gavialis gangeticus]
Psin-XP_006129195.1 PREDICTED: caspase-7-like [Pelodiscus sinensis]
For the sequences that appear misplaced, is it a failing of RD-MCL or are they misannotated in the public database? It's easy enough to test by creating a gene tree with the questionable sequence(s), a few sequences from the group it has been assigned to, and a few sequences from the group it is annotated as belonging to. The example below looks at the issue in bird_0_7, taking sequences from caspase-8 and CFLAR:
$: seqbuddy bird_casps.fa --pull_records 'Falb-XP_005049226' 'Fche-XP_014133274' 'Fper-XP_013161015' 'Gfor-XP_005415992' 'Ggal-NP_989923' 'Aaus-XP_013807673' 'Acar-XP_010167151' 'Achl-XP_009074486' 'Achr-XP_011574988' 'Avit-XP_009876193' | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree raxml
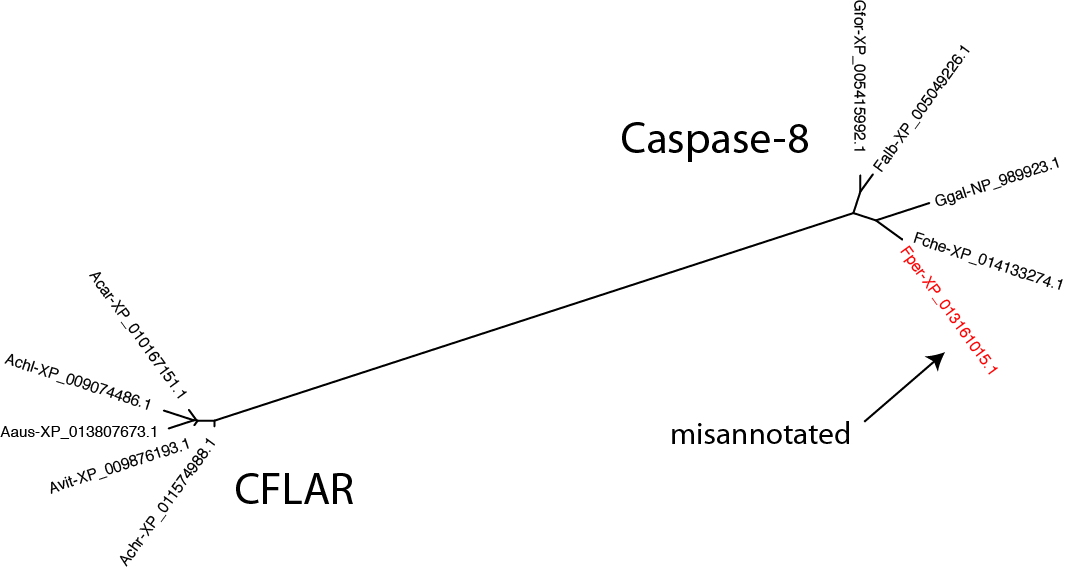
Clearly, the Falco peregrinus sequence has been assigned the wrong name in GenBank.
Similarly, to check on an uncharacterized sequence, include sequences from every cluster and determine which group it most closely groups with:
$: seqbuddy bird_casps.fa --pull_records Tgut1-XP_002191294 Tgut1-XP_012428936 Tgut1-XP_002197219 Tgut1-XP_002188318 Tgut1-XP_012430035 Tgut1-XP_002188263 Tgut1-XP_002192067 Tgut1-XP_012430417 Tgut1-XP_002190143 Tgut1-XP_002190691 Talb-XP_009970325 Talb-XP_009966940 | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree raxml

This result confirms that the uncharacterized sequence has been properly classified as caspase-3 by RD-MCL.
To generate a nice visual representation of your RD-MCL results, use the included orthogroup_polytomy_builder program. It will convert the final_clusters.txt file into a nexus formatted tree file, combining each group into a color coded polytomy.
$: orthogroup_polytomy_builder bird/final_clusters.txt > bird/bird_casps.nex
Use your favorite phylogenetic tree visualization software to open up the file (I recommend FigTree). In the tree below, I have exported the image as SVG and annotated each cluster with the appropriate name in Adobe Illustrator.
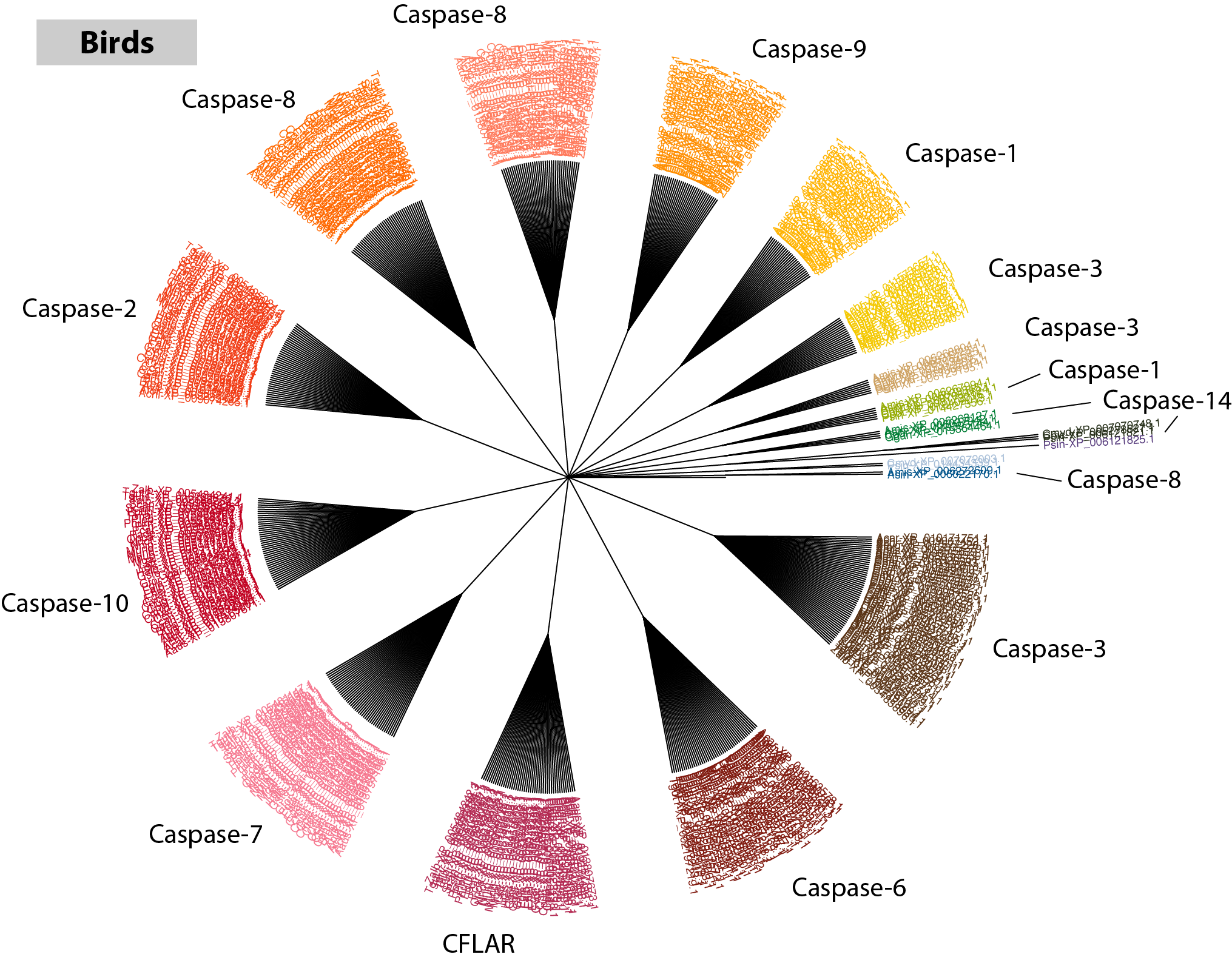
It was easy to miss while scrolling through the list of accessions and gene descriptions, but visualizing the data in tree form and annotating each cluster relative to the names pulled from RefSeq makes it clear that multiple large clusters exist for both Caspase-8 and Caspase-3. What might be the most probable explanations for this?
- Was a single large orthogroup incorrectly split up by RD-MCL?
- Do the subdivided orthogroups represent 'recent' paralogs?
- Have the genes been incorrectly assigned in the database?
All three possibilities are fairly easy to test.
Our RD-MCL result will be supported if the members of each putative orthogroup forms a monophyletic clade on a maximum likelihood tree. Below, the seqs mode and --group flag of group_by_cluster are used to pull out the Casp3 and Casp8 orthogroups, plus Casp9 as an outgroup.
$: group_by_cluster bird --mode seqs --group bird_0_0_0 bird_0_6 bird_0_7 bird_0_8 bird_0_10 bird_0_11 bird_0_15 bird_0_16 | \
seqbuddy --order_recs_by_len | \
alignbuddy --generate_alignment clustalo "--pileup" | \
phylobuddy --generate_tree fasttree -o nexus > bird/casp_3_8_9.nex

Given how cleanly separated the groups are, it's pretty safe to say that RD-MCL is performing as expected.
Note: In this example, SeqBuddy was used to order all of the incoming records by sequence length from smallest to largest, then AlignBuddy called Clustal Omega with the --pileup option. 'Pileup' iteratively builds the alignment based on the order of the sequences provided, which ensures that all sequences are anchored to the smallest common domain used to define the protein family. This is also the default behavior of RD-MCL.
Now that orthogroups are defined, the phylogenetic relationships among each of those groups can be analyzed. To do so, first create a consensus sequence for each cluster and then infer a tree from there. Again, group_by_cluster can help us out by converting each orthogroup into a consensus sequence; simply pass in the keyword cons:
$: group_by_cluster bird --mode cons | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree raxml -o nexus > bird/bird_consensus.nex

Working from consensus sequences cleans up much of the noise and greatly reduces runtime compared to inferring very large trees from hundreds of sequences. Two things pop out from the above tree: 1) The Casp8 orthogroups form a monophyletic clade, indicating that they can be considered 'recent' paralogs in birds, and 2) The Casp3 orthogroups are separated by Casp6 and Casp7, which suggests an issue with current nomenclature (see hypothesis 3, below).
Note: For this example the 'orphaned' sequences (i.e., clusters with only 1 or 2 records) in bird_0_15, bird_0_16, and bird_0_13_1 were included. We'll see later that more difficult cases can leave many orphans, so it is often helpful to set a minimum cluster size with the group_by_cluster --min_group_size flag.
Clearly, there are two distinct orthogroups in birds that have been annotated as Casp8. This may not not technically be a case of misannotation, however, because the gene duplication leading to these two orthogroups may be restricted to Archelosauria (indeed, we'll see this is the case in the next section). It's confusing to not differentiate them, but not necessarily incorrect. The two Casp3 orthogroups, on the other hand, are absolutely an annotation error. Both Casp7 and Casp6 share more recent common ancestors with the bird_0_0_0 Casp3 than with the bird_0_10/11 Casp3.
Let's first alleviate any misgivings you may have about using consensus sequence phylogenies, which are not commonly used at present. Make a tree from all the sequences in the Casp3, 6, 7 neighborhood, using one of the Casp14 clusters as an outgroup:
$: group_by_cluster bird --mode seqs --group bird_0_0_0 bird_0_1 bird_0_3 bird_0_10 bird_0_11 bird_0_14 | \
seqbuddy --order_recs_by_len | \
alignbuddy --generate_alignment clustalo "--pileup" | \
phylobuddy --generate_tree fasttree -o nexus > bird/casp_3_6_7_14.nex
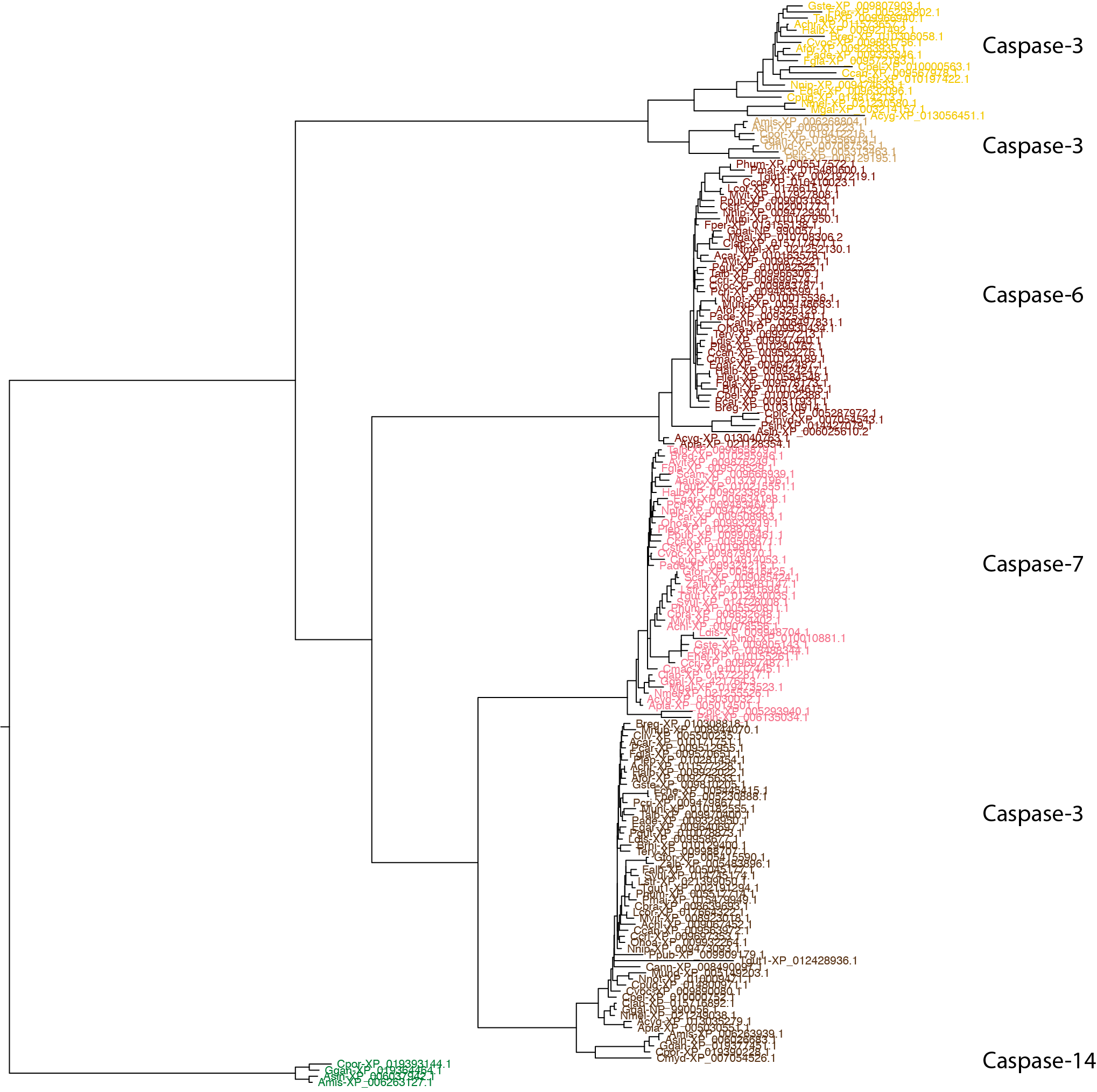
The branching pattern among orthogroups is the same (trust in the consensus!). So what might have caused such an egregious oversight in RefSeq? The 'problem' is that we're sequencing genomes at breakneck speed and there's a ton of evolution to sift through (not actually a bad problem to have, in my opinion). There isn't enough person-power at any one institution to tease apart all of the intricacies of every gene family, which is why it's now up to us in the community to perform detailed analyses on our pet proteins. But I digress — let's get back to what's up with Casp3.
The reason for this misnaming is simple: When the first bird genome was sequenced, its predicted gene models were BLASTed against reference organisms. The bird_0_10/11 orthogroup did not exist in those references, but it was still homologous to caspases. From our analysis here, we know that it is equally related to the orthogroups represented by bird_0_1 (Casp6), bird_0_0_0 (Casp3), and bird_0_3 (Casp7), but at the time, the top hit just happened to be Casp3. Therefore, it was named Casp3, even though it would have been only a marginally better hit than that for Casp6 or Casp7. The name was cemented in at this point, however, so the next time a bird_0_10/11 sequence was identified, of course its top hit was the faux-Casp3 from the first bird genome. I assure you, RefSeq is riddled with cases of misannotation like this.
The bird caspases are a very easy group to work with because the orthogroups are so cleanly separated from one another. In fact, this tends to be true across most gene families, so I highly recommend including them in your analysis. The mammals also tend to generate rather clean clusters.
$: orthogroup_polytomy_builder mammal/final_clusters.txt > mammal/mammal_casps.nex
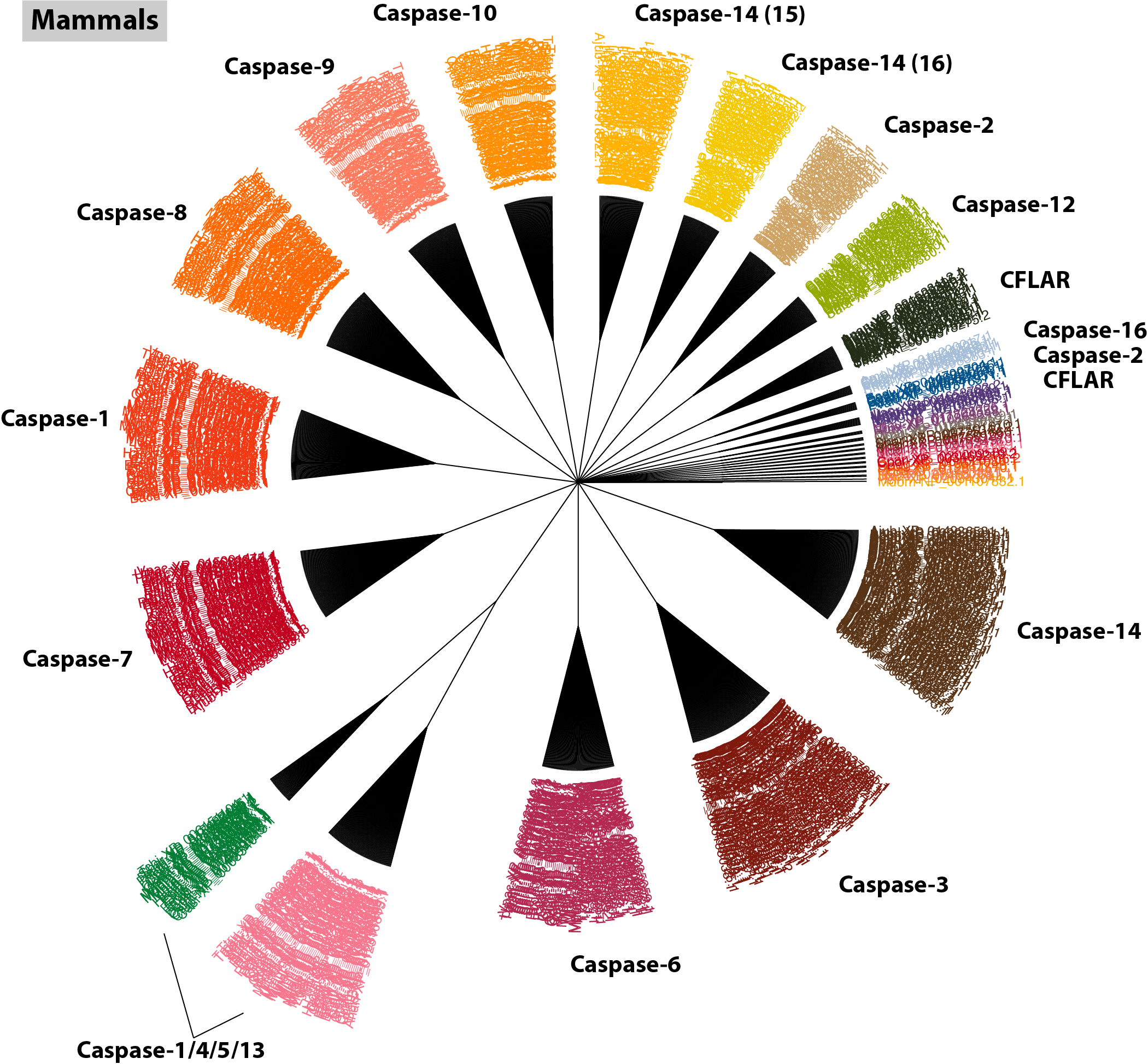
$: group_by_cluster mammal --mode cons | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree raxml -o nexus > mammal/mammal_consensus.nex

As mentioned earlier, including small clusters with orphaned sequences can clutter the tree, but keeping them in this mammal tree nicely illustrates that the method is working; all of the orphaned sequences have been pre-annotated in RefSeq and they all group closely with their larger consensus orthogroups (you can confirm this with $: group_by_cluster mammal/final_clusters.txt mammal_casps.fa). For reference, the size of each orthogroup is indicated in parentheses beside each gene name. Interestingly, this tree also supplies new information pertaining to the Casp8 orthogroups we observed in the bird analysis. Notice mamm_0_43; it contains a single orphaned record that has been annotated as Casp18. Have a quick look at its GenBank record:
$: seqbuddy refseq_casps.gb --pull_records 'NP_00110783'
LOCUS NP_001107832 471 aa linear MAM 01-MAY-2016
DEFINITION caspase-18 [Monodelphis domestica].
ACCESSION NP_001107832
VERSION NP_001107832.1
KEYWORDS RefSeq.
SOURCE Monodelphis domestica (gray short-tailed opossum)
ORGANISM Monodelphis domestica
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Metatheria; Didelphimorphia; Didelphidae; Monodelphis.
REFERENCE 1 (residues 1 to 471)
AUTHORS Eckhart,L., Ballaun,C., Hermann,M., VandeBerg,J.L., Sipos,W.,
Uthman,A., Fischer,H. and Tschachler,E.
TITLE Identification of novel mammalian caspases reveals an important role
of gene loss in shaping the human caspase repertoire
JOURNAL Mol. Biol. Evol. 25 (5), 831-841 (2008)
PUBMED 18281271
FEATURES Location/Qualifiers
CDS 1..471
/gene="CASP18"
/coded_by="NM_001114360.1:1..1416"
/db_xref="GeneID:100017025"
ORIGIN
1 mdeillqikd kltrsdlesl kflssdiipl qkqenitrsl affqalqkre lldnnsilee
61 llfligrkdi ltkqlnvnie elkrklqhsd slkispyrkl lfkiaenmts dyldsakfll
121 rrdlpqskle giktplklfi emekhgliek tnlkalkdil qslnaeclik hikeyekktk
181 dlagsavtaq leshlghlsl lsgnagqaaa plnipeesem ntlpqsmppy kmehvphgyv
241 viidnihfsn pvdvrigtek dvaalrkvfg rlqfkeeyhs nldasqlhev mkdyskrdyt
301 dqdaficcil shgkkgvvlg tdwkpvaikk llsyftanec ktlkdkpklf fiqacqngks
361 dslpevdvef dleadaicsn tthewsdifi gmatvedsla qrsgsigspy iqnlckelea
421 hcpqkkelle imtsvnskvs niiqmpefrs tlrapfifqv peesqkapig h
//
In the publication referenced in the GenBank record, Eckhart et. al. explains that "...caspase-18 originated, presumably by duplication of caspase-8 or -10, after the split of the amphibian lineage from amniotes, was further conserved in birds and [monotreme] as well as [marsupial] mammals, but was deleted in a common ancestor of all placental (eutherian) mammals."
They classify the chicken 'initiator caspase' as Casp18, which is part of bird_0_6 (Casp8), so let's see if we can support their claim and classify all of the sequences in bird_0_6 as caspase-18. To do so, create a tree from all of the orthogroups in the evolutionary neighborhood of Casp-18 from both birds and mammals: Casps-8, -10, and CFLAR, with Casp-2 as an outgroup.
$: group_by_cluster mammal --mode seqs --group mamm_0_6 mamm_0_8 mamm_0_11 mamm_0_13 mamm_0_15 mamm_0_43 > Casp18.fa
$: group_by_cluster bird --mode seqs --group bird_0_2 bird_0_4 bird_0_5 bird_0_6 bird_0_7 bird_0_15 bird_0_16 >> Casp18.fa
$: alignbuddy Casp18.fa --generate_alignment clustalo | phylobuddy --generate_tree fasttree -o nexus > Casp18.nex

The Monodelphis domestica Casp18 sequence (red arrow) sits neatly before the split between alligator/turtle Casp8 sequences (blue labels) and the Casp8 sequences represented by bird_0_6. The other two Casp8 orthogroups, mamm_0_6 and bird_0_7, form a sister clade, so I would feel very confident renaming all sequences in bird_0_6 to Casp18. Eckhart carefully assessed genomic synteny information to piece together their evolutionary story regarding Casp8/18, but we are able to arrive at the same conclusions using a much less manually intensive sequence similarity based clustering approach. Furthermore, our high coverage of placental mammals indicates that Eckhart was correct to assert that the gene was lost from the last eutherian common ancestor.
A dedicated wiki page digs further into how orphaned sequences can be utilized, but from here on we will focus on genes contained in larger, more highly conserved, orthogroups. Use the --min_size flag to strip out small clusters:
$: group_by_cluster mammal/final_clusters.txt mammal_casps.fa cons --min_size 5 | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree raxml -o nexus > mammal/mammal_consensus_min5.nex

The naming of genes in the Casp2 and CFLAF orthogroups are very consistent, but both have one large cluster and then a second smaller cluster. Let's have a look at all of these sequences to see if we can identify the cause of the splits (using Casp12 as an outgroup):
$: group_by_cluster mammal/final_clusters.txt mammal_casps.fa seqs --group mamm_0_11 mamm_0_12 mamm_0_13 mamm_0_14 mamm_0_15 | \
alignbuddy --generate_alignment clustalo | \
phylobuddy --generate_tree fasttree -o nexus > mammal/mamm_casps2_12_cflar.nex

Hmmm... Okay, the sequences in each cluster all seem to, well, cluster together on the tree. The Casp2s are both most closely related to one another, as are the CFLARs. Once again, we might be tempted to fault RD-MCL for breaking up groups that should otherwise be combined, but have a look at the actual sequence alignment for the two Casp2 groups.
$: group_by_cluster mammal/final_clusters.txt mammal_casps.fa seqs -g mamm_0_14_0 mamm_0_11 | \
alignbuddy -generate_alignment clustalo
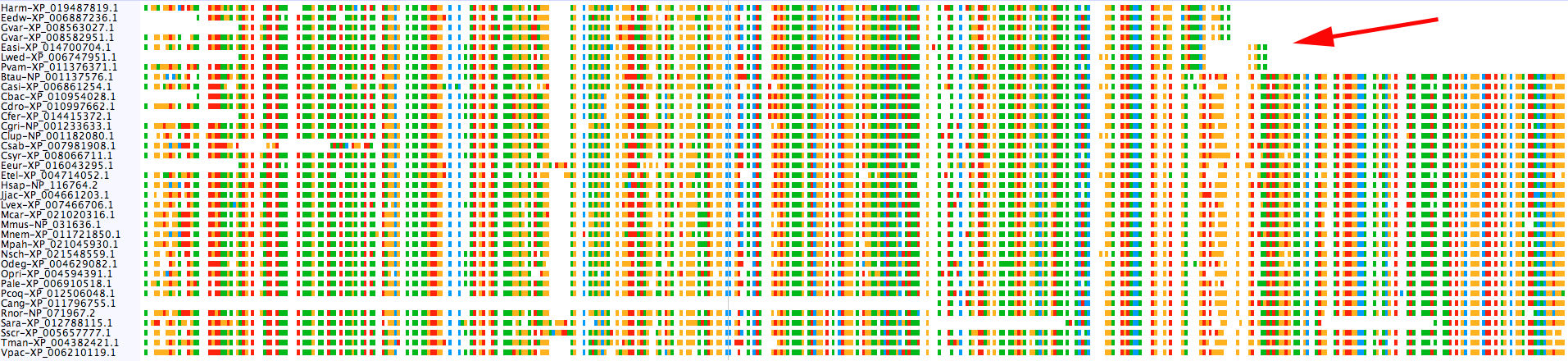
All of the sequences in mamm_0_14_0 are missing a large chunk off of their C-terminal end. The taxonomic distribution of the group does not suggest an obvious evolutionary explanation (i.e., they are scattered throughout the distribution of mamm_0_11), so my guess is an issue with genome assembly/annotation. As an example, there is a large assembly gap following the annotated Leptonychotes weddellii Casp2 gene:
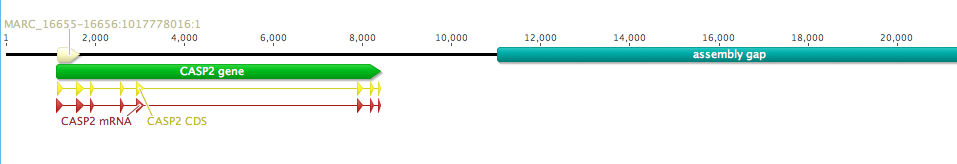
Given this obvious issue, I would personally leave these clusters separated, or even go so far as to remove the mamm_0_14_0 sequences completely from the dataset; if we suspect that the issue is technical and not real biology, there is little reason to pollute our carefully curated orthogroups with extra noise.
The CFLAR clusters do not show the same problem:
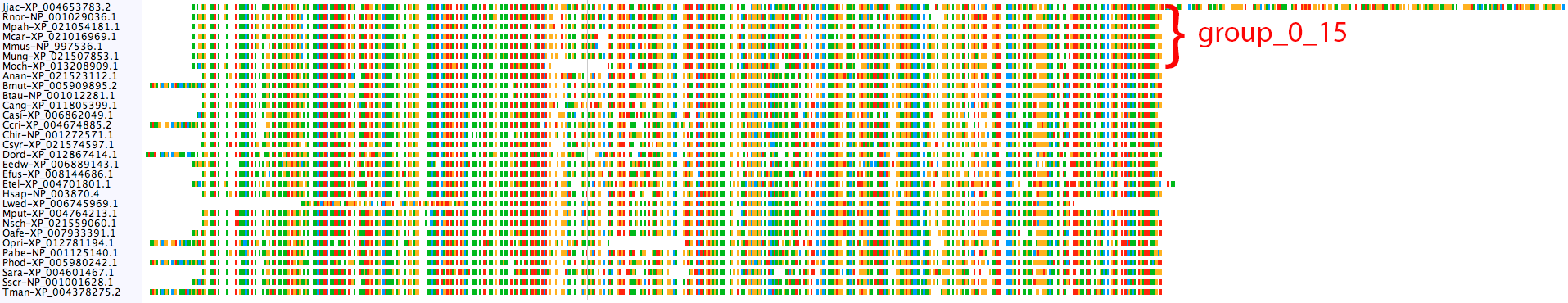
Nothing really looks amiss, except maybe the long C-terminal end of Jjac-XP_004653783.2. It's probably safe to merge mamm_0_13 and mamm_0_15 into a single orthogroup, but we can further test if it's appropriate using the merge_orthogroups tool.
$: merge_orthogroups mammal mamm_0_15
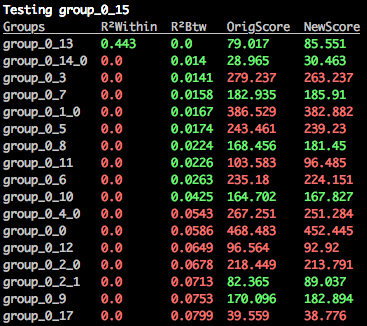
The best cluster that mamm_0_15 could be merged with has been identified as mamm_0_13, as we expected, and it's all green.
We can go ahead and merge it by re-running merge_orthogroups with the --merge flag.
$: merge_orthogroups mammal mamm_0_15 --merge mamm_0_13
You will be prompted to confirm the merge, following which the final_clusters.txt file will be updated.
BEFORE MERGE:
mamm_0_13 63.7586 Anan-XP_021523112.1 Bmut-XP_005909895.2 Btau-NP_001012281.1 Cang-XP_011805399.1 Casi-XP_006862049.1
Ccri-XP_004674885.2 Chir-NP_001272571.1 Csyr-XP_021574597.1 Dord-XP_012867414.1 Eedw-XP_006889143.1 Efus-XP_008144686.1
Etel-XP_004701801.1 Hsap-NP_003870.4 Lwed-XP_006745969.1 Mput-XP_004764213.1 Nsch-XP_021559060.1 Oafe-XP_007933391.1
Opri-XP_012781194.1 Pabe-NP_001125140.1 Phod-XP_005980242.1 Sara-XP_004601467.1 Sscr-NP_001001628.1 Tman-XP_004378275.2
mamm_0_15 14.1775 Jjac-XP_004653783.2 Mcar-XP_021016969.1 Mmus-NP_997536.1 Moch-XP_013208909.1 Mpah-XP_021054181.1
Mung-XP_021507853.1 Rnor-NP_001029036.1
AFTER MERGE:
mamm_0_13 85.551 Anan-XP_021523112.1 Bmut-XP_005909895.2 Btau-NP_001012281.1 Cang-XP_011805399.1 Casi-XP_006862049.1
Ccri-XP_004674885.2 Chir-NP_001272571.1 Csyr-XP_021574597.1 Dord-XP_012867414.1 Eedw-XP_006889143.1 Efus-XP_008144686.1
Etel-XP_004701801.1 Hsap-NP_003870.4 Jjac-XP_004653783.2 Lwed-XP_006745969.1 Mcar-XP_021016969.1 Mmus-NP_997536.1
Moch-XP_013208909.1 Mpah-XP_021054181.1 Mput-XP_004764213.1 Mung-XP_021507853.1 Nsch-XP_021559060.1 Oafe-XP_007933391.1
Opri-XP_012781194.1 Pabe-NP_001125140.1 Phod-XP_005980242.1 Rnor-NP_001029036.1 Sara-XP_004601467.1 Sscr-NP_001001628.1
Tman-XP_004378275.2
Now let's say you have a bunch of new putative caspase sequences, isolated from species with insufficient taxonomic coverage to generate high quality orthogroups on their own.
To check a new sequence that wasn't included in the original RD-MCL run, use the place_sequence program.
Example sequence: Ggar_casc.fa
>Ggar-xyz123foo Casc containing sequence from spotted garbargal of gar [Garbargal garususus]
MQTNSVHEVFEQDHITWQNRLRESKGTLADGSKNLLQSLPILGTDIRQIDALMLELMVRN
VEVQFKQQTSEADIFALSSCIFLAKGENPYVYSYDDKEKLQVVWKLLVGKNMKQLVGKYR
AGSEACPGLKDQVVIAQADIVDSAVSSANTAKVKKQPLYALKCGIARQLKPENVRGFLGK
RSNVLGSWWLSMANEFLHRVPMAVPFAEVLLVWARCAAHRNNNISHDTHAICITRMCKFP
LILLRLLFFNTDLK
$ place_sequence Ggar_casc.fa bird
Output:
Placement of sequence(s) in RD-MCL clusters
R² between clusters: 0.000 - 0.259
R² within clusters: 0.925 - 1.000
FwdScore between clusters: 9.938 - 236.542
FwdScore within clusters: 289.924 - 766.872
Testing Ggar-xyz123foo
Group Size R² 95%-CI Forward 95%-CI
bird_0_1 47 0.5124 - 0.5459 59.71 - 77.03
bird_0_3 45 0.0108 - 0.0156 5.56 - 9.06
bird_0_0_0 56 0.0019 - 0.0073 -0.72 - 4.65
bird_0_7 38 0.0075 - 0.0173 -6.44 - 2.54
bird_0_4 45 0.0096 - 0.0157 -6.46 - 2.43
bird_0_11 7 0.0010 - 0.0069 -6.76 - -3.83
bird_0_6 42 0.0131 - 0.0203 -7.49 - -2.62
bird_0_5 42 0.0157 - 0.0198 -7.68 - -6.42
bird_0_8 33 0.0098 - 0.0200 -8.66 - -6.14
bird_0_12 6 0.0092 - 0.0180 -8.89 - -7.41
bird_0_2 46 0.0192 - 0.0262 -9.21 - -5.59
bird_0_10 19 0.0072 - 0.0135 -9.44 - -5.2
bird_0_9 27 0.0105 - 0.0157 -9.48 - -6.51
This sequence is really divergent .
Still working on this tutorial
Papers using the paralogs of chicken Casp3 and Casp8 that have been annotated as such in GenBank, but make no mention of the other two (labelled as 'uncharacterized protein' and 'initiator caspase', respectively).