BioMiner is dedicated to building a data mining platform that integrates high-quality multi-omics data management, distribution and exploratory analysis.
Please access Metadata System and Indexd to get metadata and multi-omics data.
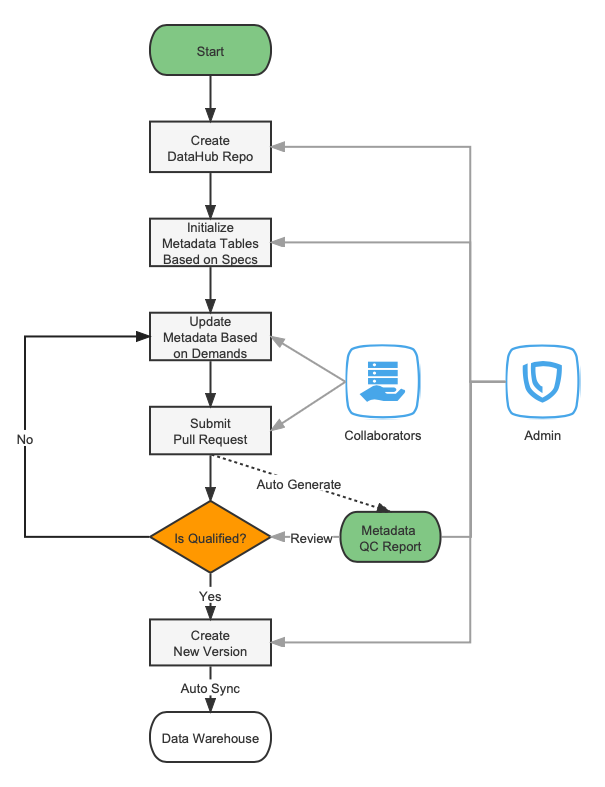
The DataHub repository is used to manage the metadata and specifications for the TCOA project.
|- .github -> Scripts for Continuous Integration and Delivery (Metadata QA & QC, Syncing the Metadata to the BioMiner System)
|- data -> Metadata Tables (Each sub directory (named with the project name) contains several metadata tables for every entity, please read the specifications for more details)
| |- FUSCC_LUAD
| | |- project/
| | |- ...
| | |- README.md
| |- FUSCC_TNBC
| |- FUSCC_CBCGA
|- docs -> Specifications and How-To
|- README.md -> Quick Start
|- CHANGELOG -> Change Log
|- LICENSE -> License File
The SEQC DataHub repository is used to manage the metadata and specifications for the SEQC project.
|- .github/ -> Scripts for Continuous Integration and Delivery (Metadata QA & QC, Syncing the Metadata to the BioMiner System)
|- data/ -> Metadata Tables (Each sub directory (named with the project name) contains several metadata tables for every entity, please read the specifications for more details)
| |- <PROJECT_NAME>_RNA/
| | |- project/
| | |- donor/
| | |- biospecimen/
| | |- reference_materials/
| | |- library/
| | |- sequencing/
| | |- datafile/
| | |- README.md
| | |- CHANGELOG
|- docs/ -> Specifications and How-To
|- README.md -> Quick Start
|- CHANGELOG -> Change Log
|- LICENSE -> License File
A centralized location for storing curated data ready for inclusion in cBioPortal.
flowchart LR
subgraph HPC [Testing on Local HPC]
publications([Publications]) --Summarized by Pipeline Developer--> scripts[Pipeline with PBS Scripts]
scripts --Evaluated by Pipeline Developer--> tested_pipeline[The Best Pipeline]
end
subgraph AliCloud [Running on AliCloud]
app[Usable and Low-cost App] --Deployed by System Developer--> platform[ClinicoOmics Web System]
end
HPC -- Pipeline >>> App --> AliCloud
HPC -. Developed by App Developer .-> AliCloud
The whole system mainly consists of the following parts:Omics Datasets' Repo, Metadata QC & QA System, Workflows & Bioinformatics Workflow Management System, Omics Data Commons(Similar to Genomics Data Commons), A Web Platform for Collaborative Multi-omic Data Visualization and Exploration(Simimar to cBioportal).
| Components | Temporary Solution | Production | Description |
|---|---|---|---|
| Omics Datasets' Repo | TCOA DataHub SEQC DataHub |
Same as the Temporary Solution | GitHub Repo, For Metadata Collaboration and Version Control |
| Metadata QC & QA System | Metabase Metadata Validator |
Same as the Temporary Solution | For Metadata QC & QA |
| Workflows & Bioinformatics Workflow Management System | ClinicoOmics | Same as the Temporary Solution | For Bioinformatics Workflow Management |
| Omics Data Commons | Metabase BioPoem |
Metabase BioPoem BioMiner Indexd |
Metabase - For Metadata Management of the Omics Datasets Biopoem - For High-speed Data Transfering (Sequencing Center -> Local HPC -> NODE -> AliCloud/Local HPC) BioMiner Indexd - A hash-based data indexing and tracking service providing globally unique identifiers. NODE/GSA/SRA - For Level 1/2 Data Files OSS - For Level3 Data Files |
| A Web Platform for Collaborative Multi-omic Data Visualization and Exploration | cBioportal cBioportal DataHub |
BioMiner Studio | cBioportal - Omics Data Exploration cBioportal DataHub - A centralized location for storing curated data ready for inclusion in cBioPortal. BioMiner Studio - Support Online Query and Exploration Analysis Modules(Similar to cBioportal, Stats and Machine Learning Modules |
graph TD
subgraph BioMiner's Management View
BioMiner -- Manage <> Similar with GDC --> Metadata[Metadata];
BioMiner -- Manage <> Similar with cBioportal --> AnalysisModules[Analysis Modules];
Metadata -.- AnalysisModules;
end
Metadata -- Contains --> Info[Patient/Sample Information, etc.];
Metadata -- File Link to --> Level1[Level1 Data - NODE/GSA/SRA];
Metadata -- File Link to --> Level3[Level3 Data - Cloud Storage];
SeqSite[Sequencing Center] -- Sync by Biopoem --> HPC[PGx HPC];
HPC -- Sync by Biopoem --> Level1[Level1 Data - NODE/GSA/SRA];
Level1 -- Transfer to Cloud Storage by BioPoem & Convert Level1 Data to Level3 Data by SeqPipe --> Level3;
AnalysisModules --> DownstreamAnalysis[Downstream Analysis];
Info -- Part of Samples' Metadata --> Dataset[New Dataset];
Level3 -- Part of Samples --> Dataset;
Dataset -- As Input Data --> DownstreamAnalysis(Mining Dataset with Several Analysis Modules on BioMiner);
DownstreamAnalysis --> Kownledges;
style Level1 fill:#00758f;
style Level3 fill:#00758f;
style Info fill:#00758f;
style Metadata fill:#dafbe1;
style AnalysisModules fill:#dafbe1;