Design discussion of the plugin
General: let's use TS for time series, DS for data series. Let's use "entity" to mean either edge or a node.
Personal comments/opinions are marked with Name:.
- The basic unit is a data series (DS). A DS consists of:
- an independent variable X (e.g. experiment condition) with N states X1, ..., XN
- one or more dependent variables Y1, .., YM (e.g. expression)
- a linking of dependent variables to an entity by a foreign key (edge, node). TS is then a specific type of DS with time as independent variable and values of type double. This nicely unites both discrete and continuous use cases.
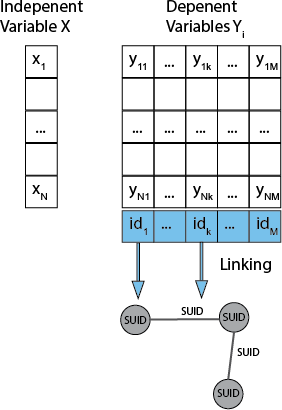
- Linking
- The independent variables of DSs can be associated with both nodes and edges, i.e. the linking, (e.g. a TS for gene expression levels associated to nodes and a TS for a predicted regulatory effect of one gene on the other associated with edges).
- The linking is based on a unique id/foreign key of the independent variables which is mapped to edge and node SUIDs (resolving the foreign key in one of the columns of edge or node table). In principle DS can also be associated with a network, but I do not see many use cases for that.
- The mapping between the dependent variables and entities must not be complete (no complete linking), i.e. part some independent variables do not need an linked entity. E.g., if mapping a DS of expression data on a pathway, there might be many nodes in the pathway that weren't part of the experiment. Scooter: do you mean that the mapping may not be complete? The way this sentence reads it sounds like you would not be allowed to have a pathway where all of the nodes have expression data.
- An independent variable could be linked to multiple entities (e.g. in a metabolic network the cofactors are often represented by multiple nodes in the network, like ATP. All these cofactor nodes should be linked to the same dependent variable). The linking is performed via an attribute which is not unique: A node/edge has exactly 1 or 0 dependent variables associated, but one dependent variable could link to multiple nodes/edges
- The special case of DS with only 1 time point must be supported, i.e. code requiring more than 1 value of the independent variable must handle the special case (for instance steady state as 1 time point timecourse).
- Multiple DS per network must be supported
- DSs have additional methods for the specific use cases. They allow for direct access to an array or doubles (double[][]) for high performance computations where necessary.
- Mixed linkings could be supported, i.e a DS can link some independent variables to nodes, other to edges (this could be handled internally by separating the different linkings of the DS)
- Some of the data will be NA values. This has to be handled.
Ideas on the actual API methods have been put at API-variants
Data series and mappings to columns are now stored with a mechanism completely separate from CyTable. Below is some discussion that led to this.
I see 3 basic reasonable ways to store TSs in Cytoscape:
- A bunch of columns in the entity table
- A a single list column in the entity table
- A separate table where each column (except a primary key) represents a time point. The entity table then has a column that references the primary key of the separate table.
I think 1 and 2 are the preferred ways, both having strengths for different use cases, but I can envision sensible uses of 3. The important point is that there is no single best way to store the data. Therefore the plugin should be flexible in data storage.
Matthias: For me 3 is by far the preferred way.
- Con 1 & 2: This pollutes the entity table (many unnecessary columns and data is mixed with other attributes). The table becomes unreadable (I often have >100 time points which does not work putting them in columns)
- Con 1 & 2: It becomes very difficult to handle multiple timeseries. Most of my data are csv tables with time as rows and columns as ids. I have many of this table which I want to load for one network (many simulations), not only one table for the network. Storing just one such "experiment/simulation set of TS" as a separate table makes things much more easy and managable. One can just create a table per csv and set the reference to the respective table when it should be used. How would one store multiple TS for the same network with approach 1 & 2 ?
- Con 2: It becomes very difficult to work with the data. One has to create an internal table for the data analysis no matter what. In addition there are issues if the time points are not at the same times for different entities or unevenly spaced, ...
- Pro 3: One has one table per experiment/simulation which only contains the time series data. One can do the data analysis/visualization easily from this table(s). There is encapsulation of the TS data.
The public TS interface defines a list of input columns in the entity table that define the data. For 1) it is all the columns involved, for 2) it is the single list column and for 3) it is the column containing the key. It is important that the input columns are part of the API so that you can check dependencies or prevent columns from deletion if they are part of a TS.
The TS further contains the definition (list) of time points. It is also has a method to provide the actual TS data (a list of values) given a CyRow. However, the way the input columns are transformed to yield a list of values for the TS is different across implementations. A nice feature of the above design is that it is possible to let plugins register their own TS implementations to define their own storage schemes for TSs if necessary (e.g. task specific database), while taking advantage of all TS support provided by the core or other plugins.
Scooter: If we're thinking of TS on top of DS, then many of the storage details are hidden at the DS layer. We could use CyTables, but we certainly don't have to. We can handle the session serialization separately. About the only real advantage using CyTables has is that if they are associated with an entity, they will be serialized in the XGMML output. I'm not sure that's a major advantage. I would store the data using an internal data structure -- still tabular, of course, but with a header that includes all of the information about the dependent variables and their associated independent variables, or sets of independent variables that go together. This would be much more efficient to work with from a computational point of view, and easier to hook up to various kinds of backends. We could certainly implement something in the API that looks like CyTable (could even implement CyTable if we wanted to), but believe me, there is lots in the CyTable implementation that works really well for associating them with entities that we might not want to deal with. So, I can think of an API that looks something like:
public List<DataTable> getAllDataTables(); // Add CyNetwork as an argument?
public DataSeries getDataSeries(DataTable dataTable, CyIdentifiable entity);
public int getDependentCount(DataSeries data);
public List<?> getIndependentData(DataSeries data);
public List<?> getDependentData(DataSeries data, int series);
public double[][] getDoubleMatrix(DataSeries data); // Throws error for discrete data
The idea is that there are two layers: a DataTable which has a bunch of metadata and the actual data series for each entity. There should be 0 or 1 independent data vectors and 1 or more dependent vectors. That way, we can easily support things like scatterplot data.
Martin: I like the idea of the DataTable/DataSeries/TimeSeries "hierarchy" - that is a nice extension. But to play devil's advocate a little - having our own data storage increases implementation effort (custom storage, custom tabular display, custom editing, ...) and sacrifices some interoperability with other plugins (e.g. I will not be able to use the data series directly with the CyNi plugin for network inference which works on CyTable). What we get is increased performance, which sounds nice, but will someone eventually perform high-performance tasks in Cytoscape? I guess that people that need heavy processing use R/numpy/Matlab/etc. anyway and then use Cytoscape only for visualisation. And the people that are not programmers and use Cytoscape as the main tool will only import data, perform some very rudimentary preprocesssing (normalization, interpolation) and then visualise. So I am not sure performance is such a big issue. Nevertheless, I think custom storage is still preferred, but I wanted to name those issues.
For more practical concerns: I would consider differentiating between returning writeable and non-writeable data (i.e. whether changes to the returned list/array will affect the stored data). This would both make the code more readable (I explicitly state that I will modify the data I got from the method) and allow us to throw an error when writing is not possible - e.g. if I construct a double[][] from a data table that is stored in a different way, I simply cannot reflect the changes automatically. Or we could just state in the contract, that the user of the API should never ever modify the data returned and use separate setXXX methods for modifying the data.
I'm also now in favour of ditching the link to CyNetwork as in the proposed API. Will save us a lot of trouble while the use cases that need to limit a DS to only a single network/subnetwork in the session seem obscure.
Getting a bit ahead of ourselves, the scheme lets you have a TS that is computed - e.g. the input columns represent a coefficients of a polynomial that you then evaluate at all the time points. Now this maybe a little clunky in some scenarios, because when the TS is computed why would you have a fixed list of time points? But in some cases, the computation depends heavily on the timepoints chosen (e.g. TS computed by numerically integrating a formula with the Euler method. Martin: I actually have a use case for this! ) so fixing the time points seems sensible and would simplify the API.
OTOH, computed TSs could be a different beast than data-based TSs and not have any fixed time points. This would slightly simplify the API at the cost of some repetitions in code and give you some nice "algebra" as in
DataTS + InterpolationMethod = ComputedTS
ComputedTS + TimePoints = DataTS
Once again, the actual implementations of computed TSs could be provided by additional plugins.
Last but not least, Computed TSs could also perform operations on multiple TSs (e.g. average) which would automatically update with changes to input data. In most cases you would probably want to actually store the computed result separately, but I see some interesting uses for automatically updating TSs.
Storage-wise, if TS definitions are stored in a separate table, plugins have enough flexibility to store anything they need to define their TS implementations (add columns for plugin-specific data etc.).
Sets of time series (e.g. repeated measurements of gene expression profiles) are handled by simply adding multiple TSs to the network. If all processing methods just have the source TS as a parameter, this should work rather nicely. It could also be possible to add another layer of indirection, i.e. to have a "virtual" TS that delegates all calls to a target TS and then change the target TS from a GUI.
Currently, metadata is stored with a custom mechanism. There is no distinction between DS connected to edges and nodes - a DS may even be connected to both edges and nodes. Some discussion, now mostly obsolete, follows.
Once we decide how to store the actual DS data, we need to know how to store the metadata (type of series, etc.). If we want to let plugins provide their own storage types/computed DSs, this needs to be public and needs to let plugins add plugin-specific metadata. I think a CyTable (similar to the "Time Series Table" in the prototype) would serve well in this context. The table would contain name and type of the DS and we would then maintain a mapping type -> provider. The provider would then read data from its specific columns in the table to construct the actual DS instance.
Should we enforce - at compile time - that a TS that is associated with edges cannot be used with nodes and vice versa?
-
Pros
- Type safety
-
Cons
- Would require messy use of generics everywhere including things like
<? extends TimeSeries<? extends CyIdentifiable>> - I may want to have a TS shared by both edges and nodes.
- I would probably need runtime checks for TS correctnes (e.g. existence of input columns) anyway.
- Since TSs need to operate on CyRow anyway and there is IMHO no direct way to get a CyIdentifiable or at least the class of the associated CyIdentifiable from the CyRow or vice versa, type safety would force part of the API calls to include both CyIdentifiable/Class and CyRow where CyRow would suffice without type safety.
- Would require messy use of generics everywhere including things like
Martin: I love type safety very much, but the cost seems quite high...
For visualisation purposes (to map to visual properties) there should be a notion of "current time" so that a value of the TSs at current time can be fed to VizMap. I see two possibilites:
- Current time is kept in the network table (all TSs have the same current time)
- Current time is a property of the TS (each TS can have a different current time)
The current time and TSs can then be made available as a formula for the equations package. We could also have an implicit hidden column for each TS showing the current time.
We could also provide the value fo the TS at current time as a virtual column...
Important things for visualization
- actual data points have to be shown (with some interpolation line if wanted), i.e. some 'o-' style
- it must be possible to see the numerical values, i.e. the table with TS data & the TS graphs at the same time. This is not working if the visualization is in the Table Panel (-> move to Results Panel).
- multiple curves must be displayed, i.e. if selecting multiple nodes/edges with time course data multiple curves have to be shown Martin: - should multiple TSs/TSs from multiple entities be shown side by side or rather over each other in a single chart? Obviously the best way is to let the user decide, but what should be our first implementation?
Large TS have to be supported efficiently. Applications are for instance
- affimetrix genome-scale time course measurement mapped on genome-scale metabolic networks (10 000 nodes, 48 time points)
- dynamical FBA simulation in genome scale networks (~4000 nodes, 500 time points)
Any thoughts?