To learn about pulsars, watch these two videos-
- https://www.youtube.com/watch?v=gjLk_72V9Bw -This is by NASA's Goddard Space Center
- https://www.youtube.com/watch?v=bKkh7viXjqs - By Astronomate.
I would highly recommend that you read this post:
| # | Mean of the integrated profile | Standard deviation of the integrated profile | Excess kurtosis of the integrated profile | Skewness of the integrated profile | Mean of the DM-SNR curve | Standard deviation of the DM-SNR curve | Excess kurtosis of the DM-SNR curve | Skewness of the DM-SNR curve | target_class |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 140.562500 | 55.683782 | -0.234571 | -0.699648 | 3.199833 | 19.110426 | 7.975532 | 74.242225 | 0 |
| 1 | 102.507812 | 58.882430 | 0.465318 | -0.515088 | 1.677258 | 14.860146 | 10.576487 | 127.393580 | 0 |
| 2 | 103.015625 | 39.341649 | 0.323328 | 1.051164 | 3.121237 | 21.744669 | 7.735822 | 63.171909 | 0 |
| 3 | 136.750000 | 57.178449 | -0.068415 | -0.636238 | 3.642977 | 20.959280 | 6.896499 | 53.593661 | 0 |
| 4 | 88.726562 | 40.672225 | 0.600866 | 1.123492 | 1.178930 | 11.468720 | 14.269573 | 252.567306 | 0 |
In this project, I decided to create a very simple Logistic Regression model (no ResNets be seen) that classifies pulsars.
The easiest way to do this would be to add this dataset on Kaggle to your notebook.See how to add data sources here on Kaggle
Otherwise, I have created a tiny script that does it for you, if you are running locally or with other Jupyter notebook providers(ie Google Colab or Binder).
from torchvision.datasets.utils import download_url
import zipfile
data_url="https://archive.ics.uci.edu/ml/machine-learning-databases/00372/HTRU2.zip"
download_url(data_url, ".")
with zipfile.ZipFile("./HTRU2.zip", 'r') as zip_ref:
zip_ref.extractall(".")
!rm -rf HTRU2.zip Readme.txt-
Create a dataframe(replace
PATH_TO_CSVwith actual path)import pandas as pd filename = "PATH_TO_CSV" df = pd.read_csv(filename)
-
Convert to numpy arrays- We need to split inputs and outputs.
Reminder- The output is the target_classimport numpy as np # Inputs # This will get everything but the target_class into a dataframe inputs_df = df.drop("target_class",axis=1) # Convert Inputs inputs_arr=inputs_df.to_numpy() # Targets-Same thing targets_df = df["target_class"] targets_arr=targets_df.to_numpy()
-
Convert to PyTorch tensors
import torch inputs=torch.from_numpy(inputs_arr).type(torch.float64)# make sure to not change the types. targets=torch.from_numpy(targets_arr).type(torch.long)
-
Create a Tensor Dataset for PyTorch
from torch.utils.data import TensorDataset dataset = TensorDataset(inputs,targets)
Now we can split the dataset into training and validation(this is a supervised model after all)
-
Set the size of the two datasets
num_rows=df.shape[0] val_percent = .1 # Controls(%) how much of the dataset to use as validation val_size = int(num_rows * val_percent) train_size = num_rows - val_size
-
Random split
from torch.utils.data import random_split torch.manual_seed(2)#Ensure that we get the same validation each time. train_ds, val_ds = random_split(dataset, (train_size, val_size)) train_ds[5]
-
I would recommend to set the batch size right about now. I am going to pick 200, but adjust this to you needs.
batch_size=200
I am giving you the option of using a GPU, but I highly do not recommend doing this as you don't need it.
from torch.utils.data import DataLoader # PyTorch data loaders train_dl = DataLoader(train_ds, batch_size, shuffle=True, num_workers=3, pin_memory=True) val_dl = DataLoader(val_ds, batch_size*2, num_workers=3, pin_memory=True) #Transfer to GPU if available def get_default_device(): #Pick GPU if available, else CPU if torch.cuda.is_available(): return torch.device('cuda') else: return torch.device('cpu') def to_device(data, device): #Move tensor(s) to chosen device if isinstance(data, (list,tuple)): return [to_device(x, device) for x in data] return data.to(device, non_blocking=True) class DeviceDataLoader(): # Wrap a dataloader to move data to a device def __init__(self, dl, device): self.dl = dl self.device = device def __iter__(self): # Yield a batch of data after moving it to device for b in self.dl: yield to_device(b, self.device) def __len__(self): #Number of batches return len(self.dl) device= get_default_device() train_dl = DeviceDataLoader(train_dl, device) val_dl = DeviceDataLoader(val_dl, device)
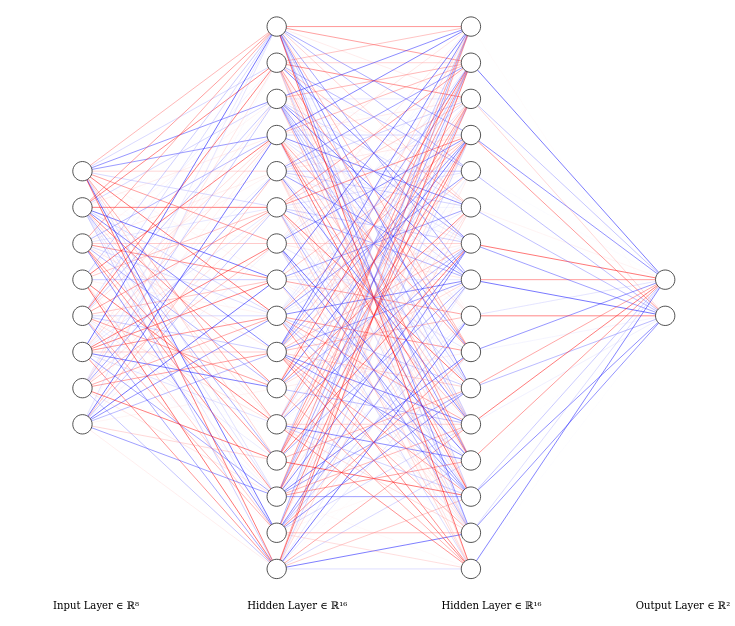
Here I decided to use a simple feed-forward neural network as in testing, it was able to reach a really good validation loss and accuracy.
- In the dataset, there are 8 inputs and one output
- The output is essentially Boolean value
- The output is 0 if it is not a pulsar or is 1 if it is a pulsar This would result in 8 neurons for the input layer and crucially two for the output layer. This is because of the Boolean output as mentioned above -one neuron will represent the probability of there being a pulsar and the other will represent the probability of signal interference
Choices:
- Maximum of 16 inner neurons in a layer- performed better than 100 neurons
- Two hidden layers.
- 10 epochs.
- Adam Optimizer
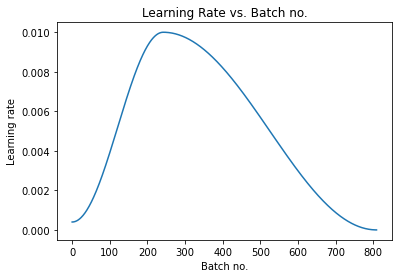
- One cycle policy
- Gradient Clipping(.1)
- Weight decay(1e-4)
- Max learning rate of .1
import torch.nn.functional as F
class HTRU2Model(nn.Module):
def __init__(self,):
super(HTRU2Model,self).__init__()
self.linear1 = nn.Linear(8, 16)
self.linear2 = nn.Linear(16, 16)
self.linear3 = nn.Linear(16, 2)
self.softmax = nn.Softmax(dim=1)
def forward(self, x):
x = x.float()# This is necessary or it would cause errors.
x = self.linear1(x)
x = F.relu(x)#I prefer to use activations functions like this, feel
x = self.linear2(x)
x = F.relu(x)
x = self.linear3(x)
x = self.linear3(x)
return x
def training_step(self, batch):
inputs, targets = batch
out = self(inputs) # Generate predictions
loss = F.cross_entropy(out, targets) # Calculate loss
return loss
def validation_step(self, batch):
inputs, targets = batch
out = self(inputs) # Generate predictions
loss = F.cross_entropy(out, targets) # Calculate loss
acc = accuracy(out, targets) # Calculate accuracy
return {'val_loss': loss.detach(), 'val_acc': acc}
def validation_epoch_end(self, outputs):
batch_losses = [x['val_loss'] for x in outputs]
epoch_loss = torch.stack(batch_losses).mean() # Combine losses
batch_accs = [x['val_acc'] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean() # Combine accuracies
return {'val_loss': epoch_loss.item(), 'val_acc': epoch_acc.item()}
def epoch_end(self, epoch, result):
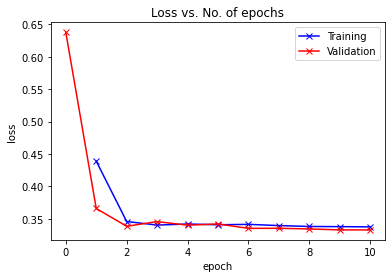
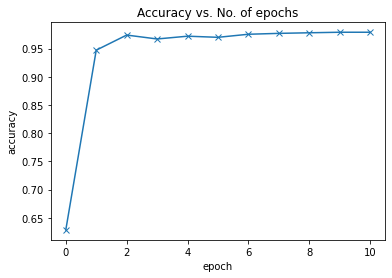
print("Epoch [{}], last_lr: {:.5f}, train_loss: {:.4f}, val_loss: {:.4f}, val_acc: {:.4f}".format(
epoch, result['lrs'][-1], result['train_loss'], result['val_loss'], result['val_acc']))I am applying the one cycle policy. Also I added a little progress bar using tqdm.
from tqdm.notebook import tqdm
def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
@torch.no_grad()
def evaluate(model, val_loader):
model.eval()
outputs = [model.validation_step(batch) for batch in val_loader]
return model.validation_epoch_end(outputs)
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group['lr']
def fit_one_cycle(epochs, max_lr, model, train_loader, val_loader,
weight_decay=0, grad_clip=None, opt_func=optim.Adam):
torch.cuda.empty_cache()
history = []
# Set up cutom optimizer with weight decay
optimizer = opt_func(model.parameters(), max_lr, weight_decay=weight_decay)
# Set up one-cycle learning rate scheduler
sched = torch.optim.lr_scheduler.OneCycleLR(optimizer, max_lr, epochs=epochs,
steps_per_epoch=len(train_loader))
for epoch in range(epochs):
# Training Phase
model.train()
train_losses = []
lrs = []
for batch in tqdm(train_loader):
loss = model.training_step(batch)
train_losses.append(loss)
loss.backward()
# Gradient clipping
if grad_clip:
nn.utils.clip_grad_value_(model.parameters(), grad_clip)
optimizer.step()
optimizer.zero_grad()
# Record & update learning rate
lrs.append(get_lr(optimizer))
sched.step()
# Validation phase
result = evaluate(model, val_loader)
result['train_loss'] = torch.stack(train_losses).mean().item()
result['lrs'] = lrs
model.epoch_end(epoch, result)
history.append(result)
return historyepochs = 10
max_lr = 0.01
grad_clip = 0.1
weight_decay = 1e-4
opt_func = torch.optim.Adamhistory = [evaluate(model, val_dl)]
historytakes ~6 seconds
%%time
history += fit_one_cycle(epochs, max_lr, model, train_dl, val_dl,
grad_clip=grad_clip,
weight_decay=weight_decay,
opt_func=opt_func)- alexlenail.me for the Neural network design program.
R. J. Lyon, B. W. Stappers, S. Cooper, J. M. Brooke, J. D. Knowles, Fifty Years of Pulsar Candidate Selection: From simple filters to a new principled real-time classification approach, Monthly Notices of the Royal Astronomical Society 459 (1), 1104-1123, DOI: 10.1093/mnras/stw656
- You can use an "image" data set called HTRU1 to achieve the same results(It also has a greater number of entry points at 60,000).
- An easier way to implement this would be to use SKLearn. This would also allow you to use KNN and SVN(and other classifier models) really easily. Check out this awesome data set on Kaggle