-
Notifications
You must be signed in to change notification settings - Fork 2
Input Data
Data input allows tab delimited .txt files to be imported.
For both metadata and data, each sample name and OTU name must be unique.
Data
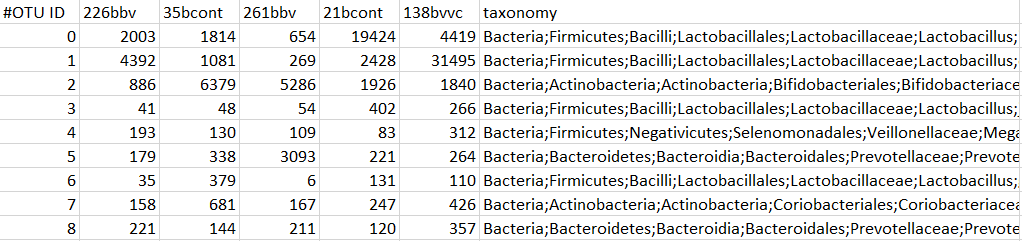
- The data file must be a tab delimited .txt (this is an option when you click 'Save as' from Excel).
- The first column must contain gene/OTU identifier.
- The first row must contain sample identifiers.
- The last column may contain taxonomic level information, but is not required. If present, it must be labelled 'taxonomy'. Taxonomy column must have at least four levels, separated by a semi colon, as shown below.
- Data table must have all blank rows removed (this may require you to check in a text editor like Notepad ++ or Atom before using the app)
Here is an example of how your data file should look:
Metadata
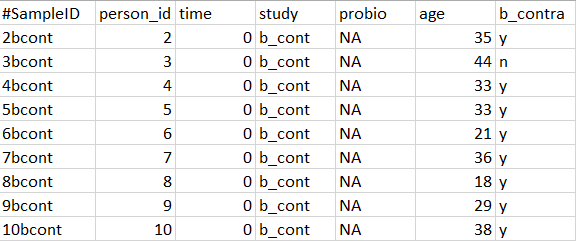
- Must be a tab delimited .txt (this is an option when you click 'Save as' from Excel).
- The first column must contain sample identifiers. The sample identifiers must be identical to the data file (not required to be in the same order).
- The first row must contain phenotypic information.
Here is an example of how your metadata file should look:
Example Data
Alternatively, you can explore omicplotR using example data provided. Click on the Example data tab on the side bar to select an example data set. The vaginal dataset has been published by Macklaim et al. (2015), and includes a metadata table, whereas the 'Selex' dataset has been published by McMurrough et al. (2014) but does not include any metadata.
After clicking the checkbox or uploading your data, click Show data and Show metadata to preview your tables, or continue by clicking 'PCA Biplots' on the navigation bar to begin visualizing your data.