-
Notifications
You must be signed in to change notification settings - Fork 2
Troubleshooting
If there are unexpected error messages or bugs, please report them by submitting an issue here. I will do my best to get in touch quickly to resolve the issue!
If you have a taxonomy file that looks like the following:
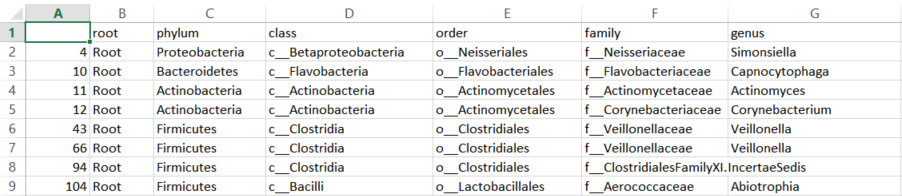
you will need to convert them so the levels are contained in one column as follows:

In R, read the table and paste the columns together as follows. Ensure the columns names match the paste line.
d <- read.table("taxon.txt", header=T, sep="\t", stringsAsFactors=F, quote = "", check.names=F, comment.char="")
taxon.short <- paste(taxon$kingdom, taxon$phylum, taxon$class, taxon$order, taxon$family, taxon$genus, sep =";")
Avoid the use of T or F as values for metadata if they do not mean TRUE or FALSE. R recognizes these as TRUE and FALSE when importing your metadata table, so any plots will be labelled as TRUE and FALSE.