Detect and select: attaching nodule centroid marker to current slice #250
Detect and select: attaching nodule centroid marker to current slice #250
Conversation
1ebaad5
to
6ee62df
Compare
interface/backend/api/views.py
Outdated
| series = candidate['case']['series'] | ||
|
|
||
| if 'files' not in series: | ||
| series['files'] = glob.glob1(series['uri'], '*.dcm') |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can you document why glob1 and not regular glob?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
|
Ran this on aws and am getting a few errors: ...
interface_1 | File "/usr/local/lib/python3.6/dist-packages/django/views/generic/base.py", line 68, in view
interface_1 | return self.dispatch(request, *args, **kwargs)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/rest_framework/views.py", line 489, in dispatch
interface_1 | response = self.handle_exception(exc)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/rest_framework/views.py", line 449, in handle_exception
interface_1 | self.raise_uncaught_exception(exc)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/rest_framework/views.py", line 486, in dispatch
interface_1 | response = handler(request, *args, **kwargs)
interface_1 | File "/app/backend/api/views.py", line 70, in get
interface_1 | ds = dicom.read_file(path, force=True)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/dicom/filereader.py", line 614, in read_file
interface_1 | force=force)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/dicom/filereader.py", line 553, in read_partial
interface_1 | stop_when=stop_when, defer_size=defer_size)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/dicom/filereader.py", line 305, in read_dataset
interface_1 | raw_data_element = next(de_gen)
interface_1 | File "/usr/local/lib/python3.6/dist-packages/dicom/filereader.py", line 219, in data_element_generator
interface_1 | value = fp_read(length)
interface_1 | MemoryError
...
vue_1 | [HPM] Proxy created: / -> http://interface:8000
vue_1 | > Starting dev server...
vue_1 | ERROR Failed to compile with 1 errors04:30:22
vue_1 |
vue_1 | error in ./src/components/open-image/OpenDICOM.vue
vue_1 |
vue_1 |
vue_1 | ✘ http://eslint.org/docs/rules/indent Expected indentation of 8 spaces but found 6
vue_1 | src/components/open-image/OpenDICOM.vue:76:9
vue_1 | this.stack.currentImageIdIndex = this.view.paths.indexOf(this.view.state)
vue_1 | ^
vue_1 |
vue_1 | ✘ http://eslint.org/docs/rules/indent Expected indentation of 8 spaces but found 6
vue_1 | src/components/open-image/OpenDICOM.vue:77:9
vue_1 | if (this.stack.currentImageIdIndex < 0) this.stack.currentImageIdIndex = 0
vue_1 | ^
vue_1 |
vue_1 |
vue_1 | ✘ 2 problems (2 errors, 0 warnings)
vue_1 |
vue_1 |
vue_1 | Errors:
vue_1 | 2 http://eslint.org/docs/rules/indent
vue_1 |
vue_1 | @ ./~/babel-loader/lib!./~/vue-loader/lib/selector.js?type=script&index=0&bustCache!./src/components/detect-and-select/CandidateList.vue 49:0-48
vue_1 | @ ./src/components/detect-and-select/CandidateList.vue
vue_1 | @ ./~/babel-loader/lib!./~/vue-loader/lib/selector.js?type=script&index=0&bustCache!./src/views/DetectAndSelect.vue
vue_1 | @ ./src/views/DetectAndSelect.vue
vue_1 | @ ./src/routes.js
vue_1 | @ ./src/main.js
vue_1 | @ multi ./build/dev-client ./src/main.js
vue_1 |
vue_1 | > Listening at http://0.0.0.0:8080
...And navigating to port |
79235b2
to
b97dd9c
Compare
|
@reubano I have fixed mentioned issues. |
|
Build seems to be failing though? :) |
|
I also noticed port 8080 was still erroring. I ran git bisect and it seems the failure is from a previous commit. Gimme a sec to find it. |
|
So according to |
|
So this is the error from travis =================================== FAILURES ===================================
____________________________ test_lung_segmentation ____________________________
self = (0002, 0001), other = (65534, 57357)
def __eq__(self, other):
# Check if comparing with another Tag object; if not, create a temp one
if not isinstance(other, BaseTag):
try:
> other = Tag(other)
/usr/local/lib/python3.6/dist-packages/dicom/tag.py:62:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
arg = (65534, 57357), arg2 = None
def Tag(arg, arg2=None):
"""General function for creating a Tag in any of the standard forms:
e.g. Tag(0x00100010), Tag(0x10,0x10), Tag((0x10, 0x10))
"""
if arg2 is not None:
arg = (arg, arg2) # act as if was passed a single tuple
> if isinstance(arg, (tuple, list)):
/usr/local/lib/python3.6/dist-packages/dicom/tag.py:21:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
signum = 14, frame = <frame object at 0x7faadaf79cf8>
def _timeout(signum, frame):
> raise TimeoutExit("Runner timeout is reached, runner is terminating.")
E app.src.tests.conftest.TimeoutExit: Runner timeout is reached, runner is terminating.
src/tests/conftest.py:96: TimeoutExit
During handling of the above exception, another exception occurred:
dicom_paths = ['/images_full/LIDC-IDRI-0001/1.3.6.1.4.1.14519.5.2.1.6279.6001.298806137288633453246975630178/1.3.6.1.4.1.14519.5.2.1...14519.5.2.1.6279.6001.490157381160200744295382098329/1.3.6.1.4.1.14519.5.2.1.6279.6001.619372068417051974713149104919']
@pytest.mark.stop_timeout
def test_lung_segmentation(dicom_paths):
"""Test whether the annotations of the LIDC images are inside the segmented lung masks.
Iterate over all local LIDC images, fetch the annotations, compute their positions within the masks and check that
at this point the lung masks are set to 255."""
for path in dicom_paths:
min_z, max_z = get_z_range(path)
directories = path.split('/')
lidc_id = directories[2]
patient_id = directories[-1]
> original, mask = save_lung_segments(path, patient_id)
src/tests/test_segmentation.py:49:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
src/preprocess/lung_segmentation.py:61: in save_lung_segments
slices = load_patient(dicom_path)
src/preprocess/lung_segmentation.py:95: in load_patient
dicom_slice = dicom.read_file(os.path.join(src_dir, s))
/usr/local/lib/python3.6/dist-packages/dicom/filereader.py:614: in read_file
force=force)
/usr/local/lib/python3.6/dist-packages/dicom/filereader.py:521: in read_partial
file_meta_dataset = _read_file_meta_info(fileobj)
/usr/local/lib/python3.6/dist-packages/dicom/filereader.py:447: in _read_file_meta_info
is_little_endian=True, stop_when=not_group2)
/usr/local/lib/python3.6/dist-packages/dicom/filereader.py:309: in read_dataset
if tag == (0xFFFE, 0xE00D):
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
self = (0002, 0001), other = (65534, 57357)
def __eq__(self, other):
# Check if comparing with another Tag object; if not, create a temp one
if not isinstance(other, BaseTag):
try:
other = Tag(other)
except:
> raise TypeError("Cannot compare Tag with non-Tag item")
E TypeError: Cannot compare Tag with non-Tag item
/usr/local/lib/python3.6/dist-packages/dicom/tag.py:64: TypeError
----------------------------- Captured stderr call -----------------------------
ERROR:root:69.xml is no valid DICOM
=============================== warnings summary ===============================
src/tests/test_endpoints.py::test_identify
/app/src/tests/../../src/preprocess/extract_lungs.py:34: RuntimeWarning: invalid value encountered in less
truncate=2.0) < intensity_th
src/tests/test_identification.py::test_identify_nodules_001
/app/src/tests/../../src/preprocess/extract_lungs.py:34: RuntimeWarning: invalid value encountered in less
truncate=2.0) < intensity_th
src/tests/test_identification.py::test_identify_nodules_003
/app/src/tests/../../src/preprocess/extract_lungs.py:34: RuntimeWarning: invalid value encountered in less
truncate=2.0) < intensity_th
-- Docs: http://doc.pytest.org/en/latest/warnings.html
========= 1 failed, 47 passed, 4 xfailed, 3 warnings in 820.90 seconds =========
The command "sh tests/test_docker.sh" exited with 1.
Done. Your build exited with 1.I also rebuilt it, and port |
…t slice, added slice index to saving properties
b97dd9c
to
3fd27e9
Compare
|
@reubano, @lamby PR has been fixed by rebasing it of top of last commit that actually fixed the issue.
This is because of the Router Guard(#239) implementation :) You should click the green bar to make it let you access the next screen: |
|
This is because of bad dummy data. Unfortunately #145 is still not closed, so the proper way to start the case doesn't work. |
|
Hmmm, how did you get your screenshots then? The image shows up for me on the master branch. |
|
@reubano this will give you correct data: |
|
Thanks, that worked! But the image only appears after moving the slider to a new slice. |
|
That's because dummy candidates are located on random and often missing slices. Opening a candidate will try to open that slice. |
|
Gotcha. Nice work! Well, LGTM. |

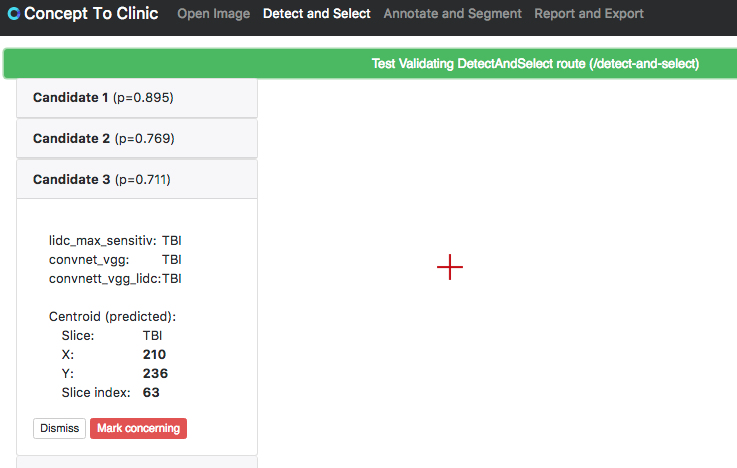
Implemented slice switching for candidate list, and a way for user to see when slice differs and attach the marker to current slice.
Description
Following features has been implemented:
On back-end I have added a method for loading candidates with cases and series. Series object will also have list of DICOM files in the folder saved in
uriproperty.Reference to official issue
It's an improvement of #148
Screenshot:
full size image
CLA