At first scZCL is a breif R package for large scale data(large DGE) from scZCL online function Zebrafish Cell Landscape ,to alleviate burdens of our main Server.
Now we add a UI for visulizing the scHCL reuslt.
#This require devtools
install.packages('devtools')
library(devtools)
# scZCL requires ggplot2/reshape2/plotly/shiny/shinythemes/shiny
install_github("ggjlab/scZCL")library(scZCL)
# zcl_caudalfin is an example expression matrix from ZCL project.
> data(zcl_caudalfin)
> dim(zcl_caudalfin)
[1] 2000 380
# 2000 genes expression value of 380 cells
# scZCL has two parameters , single cell expression matrix(scdata) and
# the number of most similar cell types
> zcl_result <- scZCL(scdata = zcl_caudalfin, numbers_plot = 3)The return of scZCL() is a list which contains 4 parts.
- cors_matrix: Pearson correlation coefficient matrix of each cell and cell type.
- top_cors: equals to numbers_plot
- scHCL: the most relevant cell type for each query cell
- scHCL_probility: the top n relevant cell types for each query cell
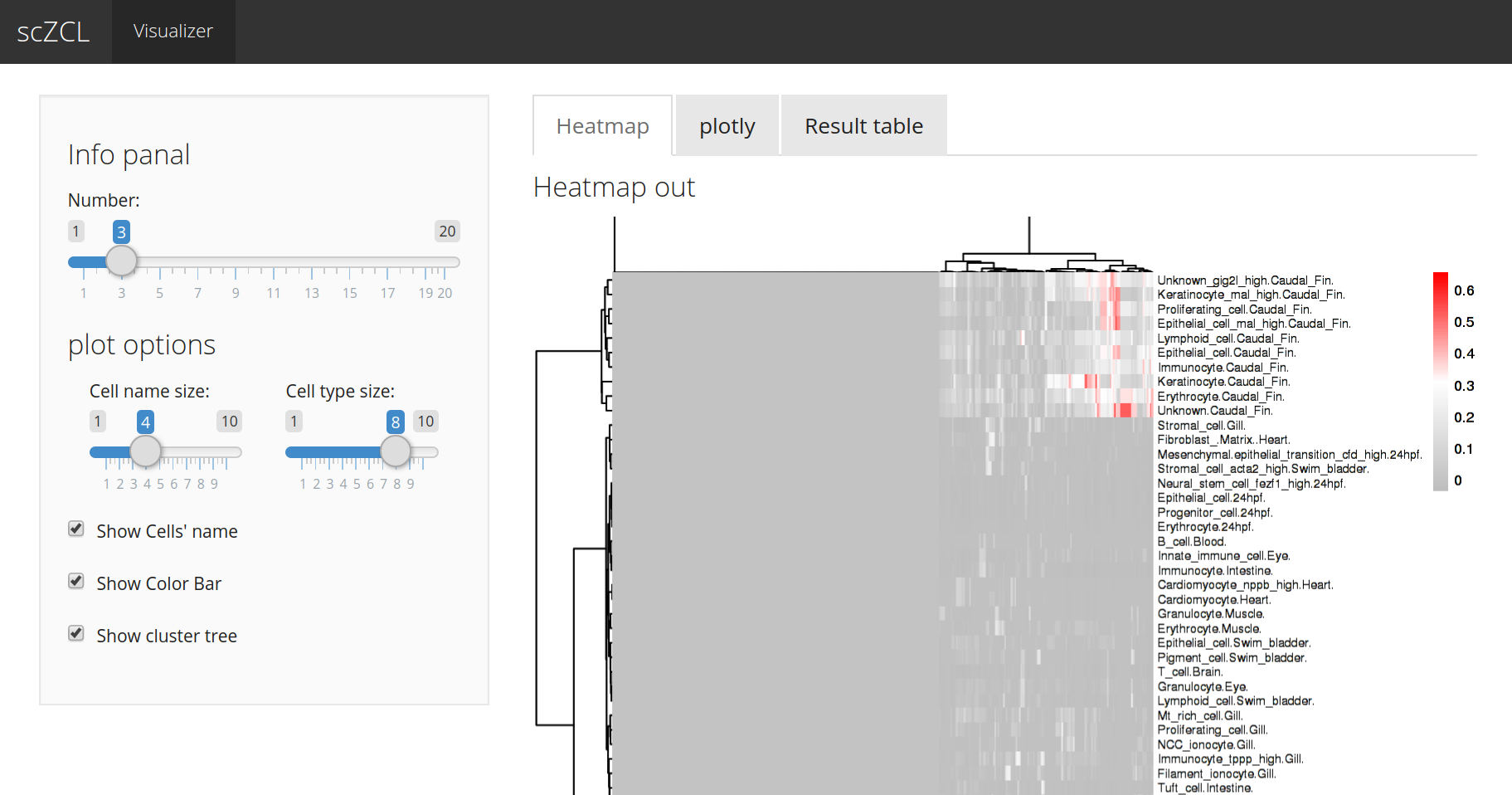
# open shiny for visualize result for scZCL
scZCL_vis(zcl_result)scZCL_vis() provides a bref function for visualizing and downloading of scZCL results