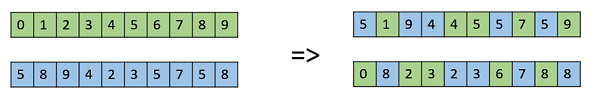This is a genetic Algorithm Library written in c++. Functions for mutation, selection, crossover and evaluation are written and can be used with any data type or any object type of your preference. If you need something more specific it is also possible to use user-defined functions.
- Swap mutation
In swap mutation, we select two positions on the chromosome at random, and interchange the values. This is common in permutation based encodings.
- Scramble Mutation
Scramble mutation is also popular with permutation representations. In this, from the entire chromosome, a subset of genes is chosen and their values are scrambled or shuffled randomly.
- Inverse Mutation
In inversion mutation, we select a subset of genes like in scramble mutation, but instead of shuffling the subset, we merely invert the entire string in the subset.
- Roulette Wheel Selection
In a roulette wheel selection, the circular wheel is divided according to the fitness of each gene. A fixed point is chosen on the wheel circumference as shown and the wheel is rotated. The region of the wheel which comes in front of the fixed point is chosen as the parent. For the second parent, the same process is repeated. Implementation is quite different but the result is identical.
- Stochastic Universal Sampling
Stochastic Universal Sampling is quite similar to Roulette wheel selection, however instead of having just one fixed point, we have multiple fixed points. Therefore, all the parents are chosen in just one spin of the wheel. Also, such a setup encourages the highly fit individuals to be chosen at least once.
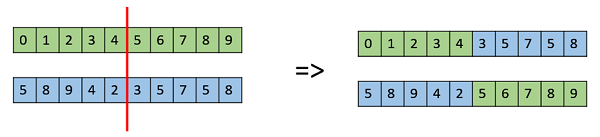
- One Point Crossover
In this one-point crossover, a random crossover point is selected and the tails of its two parents are swapped to get new off-springs.
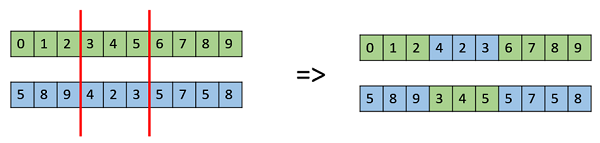
- Multi Point Crossover
Multi point crossover is a generalization of the one-point crossover wherein alternating segments are swapped to get new off-springs.
- Uniform Crossover
In a uniform crossover, we don’t divide the chromosome into segments, rather we treat each gene separately. In this, we essentially flip a coin for each chromosome to decide whether or not it’ll be included in the off-spring. We can also bias the coin to one parent, to have more genetic material in the child from that parent.
-
To create a random population of genes with char chromosomes.
-
To create a random population of genes with int chromosomes.
I'll explain this with a sample problem. Let's guess a target string from a population of random strings.
- Import necessary Libraries
#include "gencpp.h"
#include "util.h"
#include "mutation.h"
#include "selection.h"
#include "crossover.h"
- Create the fitness function which suits your needs
// How much the input string is similar to the target string
double f(vector<char> a) {
double r = 0;
string t = "target";
for (int i = 0; i < (int)a.size(); i++) r += (double)(t[i] == a[i]);
return r;
}
- Create the initial generation
vector<vector<char>> initPop = util::generateCharGenes(8,100);
- Evaluate by specifying necessary functions.
pair<vector<char>, double> best = gencpp::evaluate<char>(initPop, 100, 0.1, f,
mutation::scramble_mutate, crossover::uniform_crossover, selection::roulette_wheel_selection);
Read function descriptions for more info.