-
Notifications
You must be signed in to change notification settings - Fork 176
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Thoughts on combining 1d and 1d2 nanopore reads? #659
Comments
|
This isn't a large assembly, so run all three variations (1d, 1d2, 1d+1d2). I expect (based on gut feeling instead of actual data) that the 1d reads will dominate the correction process, so any gain from adding 1d2 will be slight - base call accuracy in the reads might be slightly better. Coverage and read length of the 1d reads could make this the best assembly, but bacteria generally assemble well, so the difference could end up just being base call accuracy, in which case 1d2 reads should help. In other words, fame and glory is yours to be had! Run the experiment and report the results! Check the FAQ for 'low coverage settings' which might help 1d2 (and add a fourth assembly to the mix). |
|
Something else you could try is running just the correction step on the 1D reads, then combining those reads with the 1D^2 reads as nanopore-corrected sequences in a standard assembly. |
|
Thanks for helping out a newbie! I ran the four assemblies that @brianwalenz described (still setting up @gringer 's suggestion). Unfortunately, this organism does not have a trust-worthy reference. In the absence of a trust-worthy reference, how would you recommend I evaluate the assemblies? 1d only assembly: 4 contigs, 1195 unassembled sequences, 8 unitigs Thanks for any suggestions! |
|
Yes, that's always tough without a good reference. You could try metaQUAST which will try to find a good reference(s) for your assembly. You could also try BUSCO to check gene completeness. It seems, as @brianwalenz predicted the 1d2 are not really helping the assembly continuity (adding them didn't change the 4 contigs you get from the 1d only asm) but it is possible they improved the consensus which the above analysis should show (by increasing BUSCO genes or improving alignment accuracy). |
|
What do the contig and unitig genome graphs look like? You can load them up in Bandage via |
|
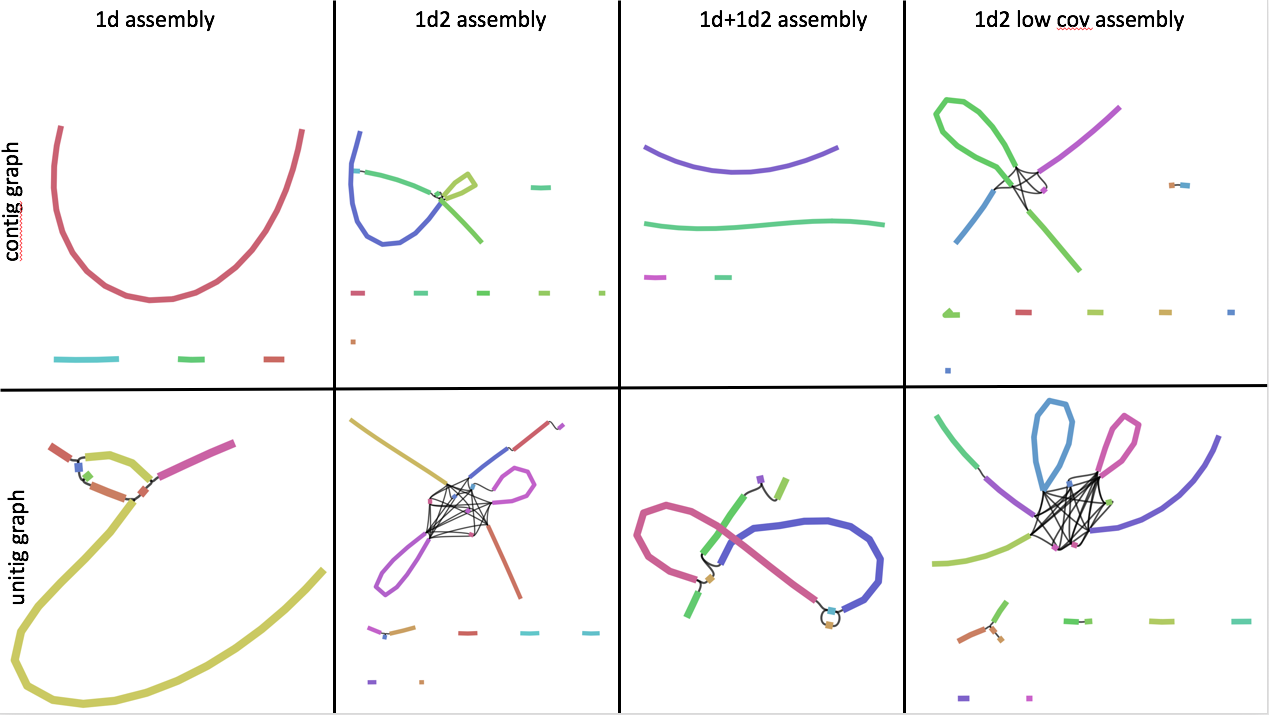
Thanks for the suggestion, @gringer ! I loaded the two graphs into Bandage for one of the assemblies. I am new to analyzing assembly graphs with Bandage -- what should I look for? Thanks! Contigs graph Unitig graph |
|
@rrwick is probably a better person for interpreting assembly graphs, but I think you're right that 1D-only and 1D+1D^2 are the least complicated (and therefore probably the closest to the truth). Neither of the graphs are showing a properly circular connection, which seems a little odd to me. My guess from the unitig graph, showing every unitig connected, is that this is likely to be a single chromosome with a bit of population variation. Were the 1D and 1D^2 from the same DNA extraction, or different samples? If different, I'd go with the 1D assembly. You could also do a genome comparison with LAST to see how different the assemblies are. |
|
I'm not too good at interpreting Canu assembly graphs because I don't have a deep understanding of Canu's inner logic. That being said, I'm inclined to agree with @gringer. If you're keen to experiment more and want the best possible assembly, here are a few suggestions that jump to mind (biased toward my own tools, because that's what I know best 😄):
Assuming your bug has a circular chromosome, you're hoping to see something like this: Or if your bug has plasmids, maybe something like this: Linear chromosome/plasmids are possible in bacterial, in which case you may not get loops like that. But I've found that linear contigs are far more often caused by incomplete assemblies than genuine linear replicons. Best of luck! |
|
Often what prevents circularization is variation in the "clonal" sample or low identity reads on the ends. How big are these contigs? It looks like the 1d2 reads split the largest contig into two. Are any of them chromosome sized? What's the output in the sizes files in unitig unitigging/4-unitig get folder? You could try manually looking for circulization as in issue #663. You could also try Canu tip, especially if you're using a version before 1.6. |
|
Closing, hopefully enough info in the comments to pick an assembly. |
|
Thanks for wrapping this up, @skoren I've been slow on this, but I did a more or less scientific comparison using different assemblers and filtering strategies, and I figured I'd post the results here for posterity. For the following results, I used only the 1d reads. Also, I found out that my sequences came from a bacterium that has a linear chromosome (a strep). So getting a circular chromosome is not a high priority anymore. I used all default command line options, unless specified in parentheses. I'm using Canu 1.6 (with For Filtlong, I tried a more lenient approach using the following options:
I also tried a more aggressive approach using these options:
As I mentioned, I don't have a good reference for this species, but based on the results below, unicycler together with the lenient filtering strategy seems to have done pretty well (@rrwick !). I'm planning on adding some more data to this experiment (other bugs, etc) and I will also try to get my hand on some data where I have a better reference. Thanks for the thoughtful discussions, everyone, and I very much appreciate all the help! |






Hi all,
I have 1d and 1d2 nanopore reads for a GC-rich microbial genome. Does anyone have experience combining these data? Is it worth it?
Thanks for any thoughts!
Edited to add:
1d read stats:
1d2 read stats:
The text was updated successfully, but these errors were encountered: