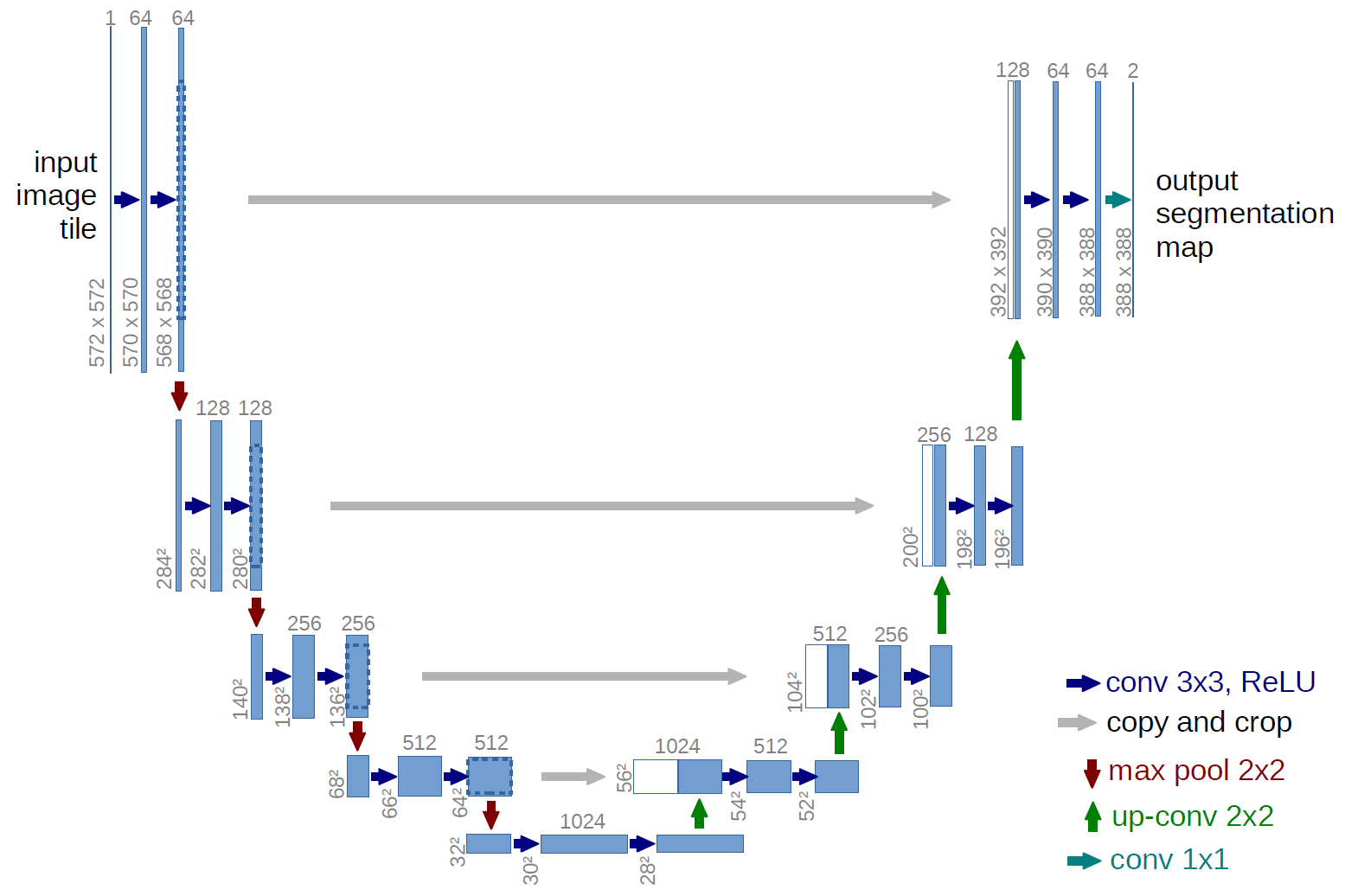
This is a implementation of U-Net for semantic segmentation using Tensorflow.
The project requires the following libraries to be installed:
- Tensorflow 2.x
- numpy
- cv2
- sklearn
- argparse
In order to train a custom dataset with U-Net, you need to have labelled data. You can either manually prepare your dataset or use Roboflow to label, prepare and host your custom data automatically in semantic segmentation mask format.
For more informations on how to prepare your dataset, see the Roboblow tutorial.
Using Roboflow to prepare your dataset Roboflow is a platform that provides tools to label and preprocess your data. Follow these steps to prepare your dataset using Roboflow:
- Sign up or log in to Roboflow and create a new project indicating "Semantic Segmentation" type. 2.Upload your images to the project. Images should be in JPEG or PNG format.
- Annotate your images. Roboflow supports various annotation formats, including bounding boxes, polygons and keypoints. For U-Net, choose "Semantic Segmentation" as the export format. This will create a mask image for each annotated image, containing the segmentation information in grayscale.
- Apply preprocessing options, if needed. Roboflow provides options such as resizing, rotation and augmentation to prepare your images and masks for training.
- Export your dataset. Roboflow will create a zip file containing your images, masks and a YAML file with information about your dataset.
Regardless of whether you use Roboflow or prepare your dataset manually, the folder structure and file types should follow these guidelines:
- Images should be stored in a folder named "images".
- Masks should be stored in a folder named "masks".
- Each image should have a corresponding mask file with the same name and ".png" extension.
- The mask files should contain the segmentation information in grayscale.
Here's an example of the contents of the "images" and "masks" folders:
├── dataset/
│ ├── images/
│ │ ├── image1.jpg
│ │ ├── image2.jpg
│ │ └── ...
│ ├── masks/
│ │ ├── image1.png
│ │ ├── image2.png
│ │ └── ...
To train a model, run the following command:
python train.py --data <data_folder> --csv <csv_output> --model <model_output> --epochs <epochs> --batch-size <batch_size> --img_ext <image_extension> --mask_ext <mask_extension> --imgsz <image_size>
You can also specify the following arguments:
--data: Data folder name. Default: data--csv: CSV to output name. Default: results_unet_train.csv--model: Model to output name. Default: model.h5--epochs: Number of epochs for training. Default: 100--batch-size: Batch size for training. Default: 2--img_ext: Image extension. Default: .jpg--mask_ext: Masks extension. Default: .png--imgsz: Image size for inference. Default: [304, 3072]
The model use a U-Net architecture with the following structure:
- Input layer: (304, 3072, 3)
- Convolutional layer 1: 32 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 2: 32 filters, kernel size 3x3, stride 1, padding 1
- Max pooling layer: kernel size 2x2, stride 2
- Convolutional layer 3: 64 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 4: 64 filters, kernel size 3x3, stride 1, padding 1
- Max pooling layer: kernel size 2x2, stride 2
- Convolutional layer 5: 128 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 6: 128 filters, kernel size 3x3, stride 1, padding 1
- Max pooling layer: kernel size 2x2, stride 2
- Convolutional layer 7: 256 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 8: 256 filters, kernel size 3x3, stride 1, padding 1
- Max pooling layer: kernel size 2x2, stride 2
- Convolutional layer 9: 512 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 10: 512 filters, kernel size 3x3, stride 1, padding 1
- Max pooling layer: kernel size 2x2, stride 2
- Convolutional layer 11: 1024 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 12: 1024 filters, kernel size 3x3, stride 1, padding 1
- Up sampling layer: size 2x2, stride 2
- Concatenation layer
- Convolutional layer 13: 512 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 14: 512 filters, kernel size 3x3, stride 1, padding 1
- Up sampling layer: size 2x2, stride 2
- Concatenation layer
- Convolutional layer 15: 256 filters, kernel size 3x3, stride 1, padding 1
- Convolutional layer 16: 256 filters, kernel size 3x3, stride 1, padding