New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
WIP: Dont center points in head coords #5085
Conversation
|
Conceptually: yes, sounds like a no-brainer. |
This is the right thing to do, yes.
This looks like the result of clustering (?), which should find sensors all with the same sign. So this is not surprising to me. |
|
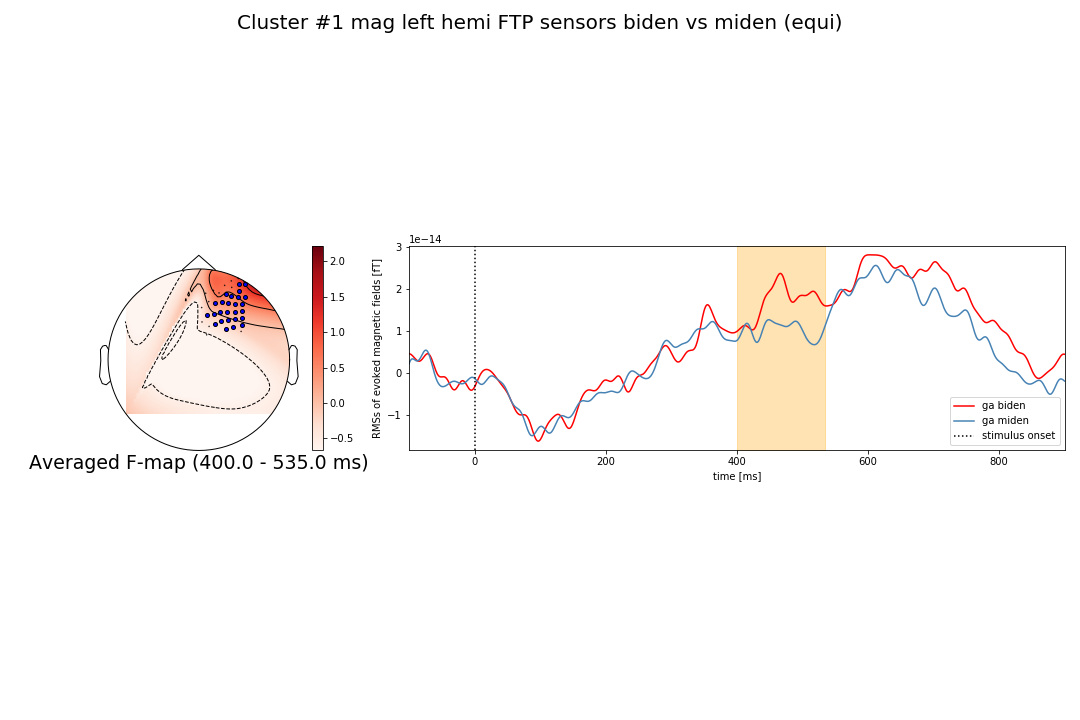
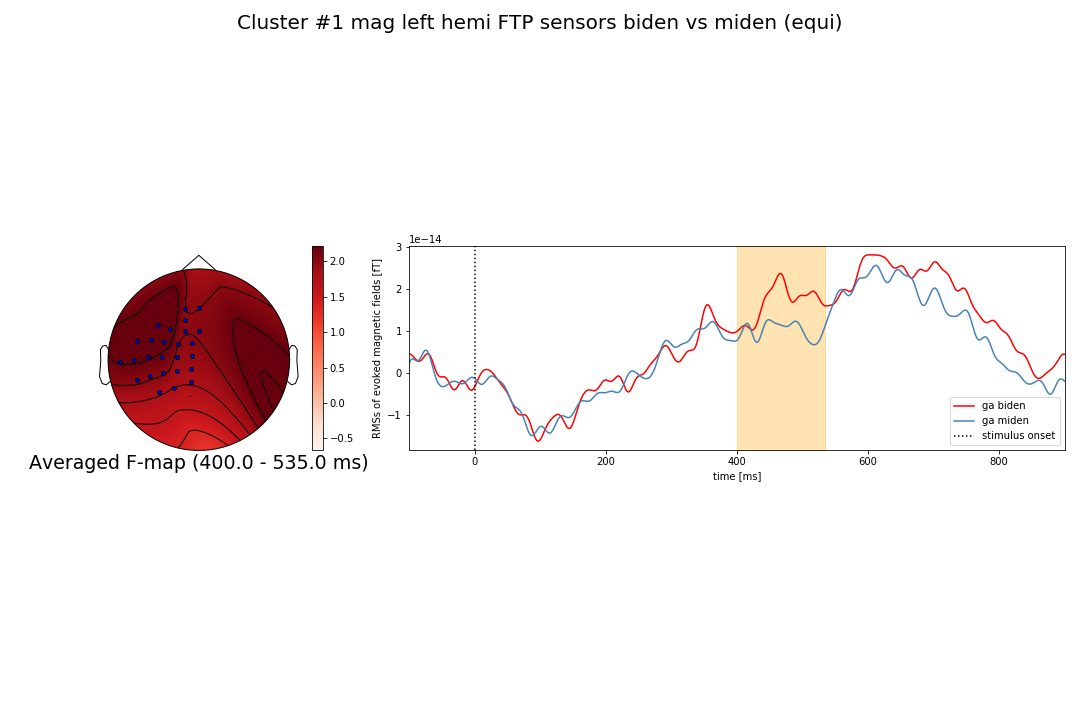
Thank you a lot larsoner. Yes I'm using (spatio_temporal_cluster_1samp_test) for permutations but since my selection of magnetometers is restricted to left sensors only: ['Left-temporal', 'Left-parietal', 'Left-frontal'], shouldn't the topography be similar to what you showed in Figure 2 (upper one) when you selected only left sensors and used head_pos (topography in the left hemisphere only)? |
|
That is what is in your second figure, no? |
|
... at least the "sensor dots" are in the left hemi I mean. The fact that the red/contours extend into the right hemisphere is a bug / limitation we will fix here. |
|
I meant the red color extending to the right hemisphere. Anyway thank you for your help, |
|
@RashaHyder why are you trying to plot a topomap for a subset of channels ..? |
|
@jona-sassenhagen |
|
I think the interpolated maps are not a good idea here. They'll inherently be extremely distorted. If at all, you'd want to interpolate over all sensors and mask for those you're actually plotting. |
|
@jona-sassenhagen tank you very much for your suggestion. I was actually concerned about a possible distortion after I saw the uni color topography in figure_2 ! |
|
For a non-interpolated legend, you can use |
|
Great! Thank you a lot I really appreciate your help. |
Agreed. The fix I have in mind will take whatever set of sensors you are using and only interpolate some small distance away from them (probably set by the distance to the next-nearest point or so). This should work regardless of whether or not channels have been picked. |
|
@larsoner an easy option would be giving the option of turning off interpolation and just plotting big colored discs. |
|
It probably makes sense to interpolate between the sensors in most use cases, no? I worry that the "disc" idea would look a bit bad in most cases due to the gaps. I imagine something like a voronoi plot would look better. But either way, this should be a new mode we can use in all circumstances, and thus (sufficiently) orthogonal to this PR. |
Yes. The zero interpolation would be for the remaining few cases :) The Voroni option will fail for sparse discontinuous channel choices. No-interpolation will never be misleading in the way interpolation methods are in that case. |
|
@jona-sassenhagen I am looking at implementing this functionality and from looking at the code |
|
Ok, will do today. |
|
To make reviewing easier, I restated the question in PR form: #5140 :) |
|
Closing this for the proposal in #5471. Basically from talking to @agramfort it probably makes the most sense for MEG topomaps to be in device coords (maybe with a head pos + rotation actually shown), and EEG topomaps in head coords. |
So far:
dev_head_tto MEG intopomap_coordsto get correct Head<->sensor relationshipdev_head_tapplication optional?montage.plotandplot_sensors)Closes #3987.
Running this code:
Gives (ignore the broad smearing plotting "artifact" due to channel subselection in the second one, we can fix this eventually), note that the subselected channels stay put, and that the sensors are rotated relative to the head (in accordance with
dev_head_tfor this dataset):Omitting the
head_posargument (which basically means "center and scale to fit the circle") the second plot gives the sort of thing that has been confusing people in #3987 and elsewhere:Assuming we have geometry information properly in the head coordinate frame (and I would argue we should make people corrrect theirs if they do not), this is incorrect. I'm thinking that we should change the default for
head_posto this (or similar) scheme. Thoughts @jona-sassenhagen ?