New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
WIP: Fix topomap plotting for EEG #5472
Conversation
|
Sounds like a good plan. What about #5269 - should we compute the locations ourselves so we have exactly one location per label? Otherwise, we should make sure that whenever we talk about a specific location, e.g. C3, its location should be the same in all our montages. |
|
I think that's an orthogonal issue, let's discuss that over in #5269 |
|
The question whether to compute the locations or whether to use a "master montage" from which all montages are derived is orthogonal, but I'd still add the point "ensure all our montages actually use the same underlying locations" to your to do list. |
|
That seems like an issue that can and should be tackled separately, this PR will be annoying enough to review and test as is |
|
OK, then let's not forget about this and discuss in #5269. |
|
Too risky to rush to finish this for 0.17, better to make it the first thing to work on in 0.18 to make sure we have months to catch and fix bugs |
|
I've editted the TODO list to remove the fix that is included in #5754. |
Regarding this second point : maybe this should be considered during |
|
as a reminder the idea of a refactor is to read from montage file a dig
object that has all necessary information (coordinate system, units etc.)
so yes it's part of the puzzle
|
|
regarding:
I have checked all the montages shipped with mne, and:
Here are all the results: |
I was recently reading through the topomap code and my thoughts went back to this issue.
All this relates to how channels are projected to 2d for EEG currently. I did not check how or whether it differes for MEG. It also makes the most sense if generic montage is used, not digitized channel positions. In the case of digitized positions the head shape would not be spherical so the current way channels are projected to 2d may look weird. But anyway, for the proposed fix to work for digitization, only the head center would have to be defined. |
|
(btw - I meant that radius is not used in |
Yes for plotting perhaps, but we still want a way of controlling it e.g. for source localization / coreg with surrogate MRI (like |
|
I just got triggered by the |
|
@larsoner |
|
@larsoner |
|
to move forward here I would suggest the following:
- open a PR with an example that shows all the things that are broken or
could be a bit improved (plot topos with different options, caps etc.)
- change the mne behavior and report the updated example we can look at to
see that things are actually better now.
|
|
@agramfort Ok, I will! |
|
I'll close this but others feel free to take over! |
Based on this and how many bugs there are, I think we should get rid of this So I think we should build off of what you did @mmagnuski, specifically:
I have made a gist containing what I think are all of the bad behaviors: https://gist.github.com/larsoner/f30e6b3b2f4e7c7979abbd367ea7ee54 My plan of action is to take @mmagnuski's code and:
Let me know if anyone sees problems with this plan. |
|
sounds like a great plan !
… |
|
@larsoner |
|
I am all in favor of throwing out |
|
@mmagnuski I think I have it almost working after a lot of refactoring. Will push here shortly then you can try it this weekend |
|
Okay @mmagnuski that's all I have time for, feel free to try it. If there are obvious bugs feel free to push commits to fix them. |
|
Argh push force then reopen is problematic, will open a new PR |
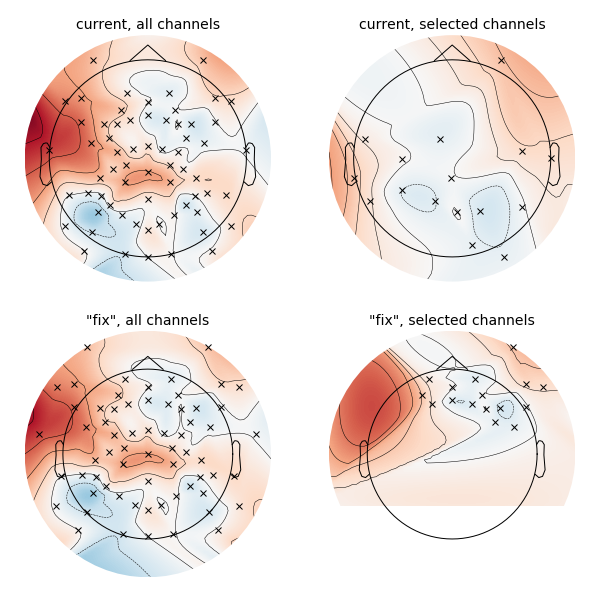
Closes #3987
Closes #4880
Closes #5190
Closes #5472
Closes #6304
Todo:
plot_sensorsbe in physical coordinatestransforminmontage,outlines, ...plot_topomap,plot_sensors,montage.plotexamples viacircle fullcc @jona-sassenhagen @sappelhoff @cbrnr you might be interested in this plan.
After this, due to using physical units for the head, #5471 should be simpler.