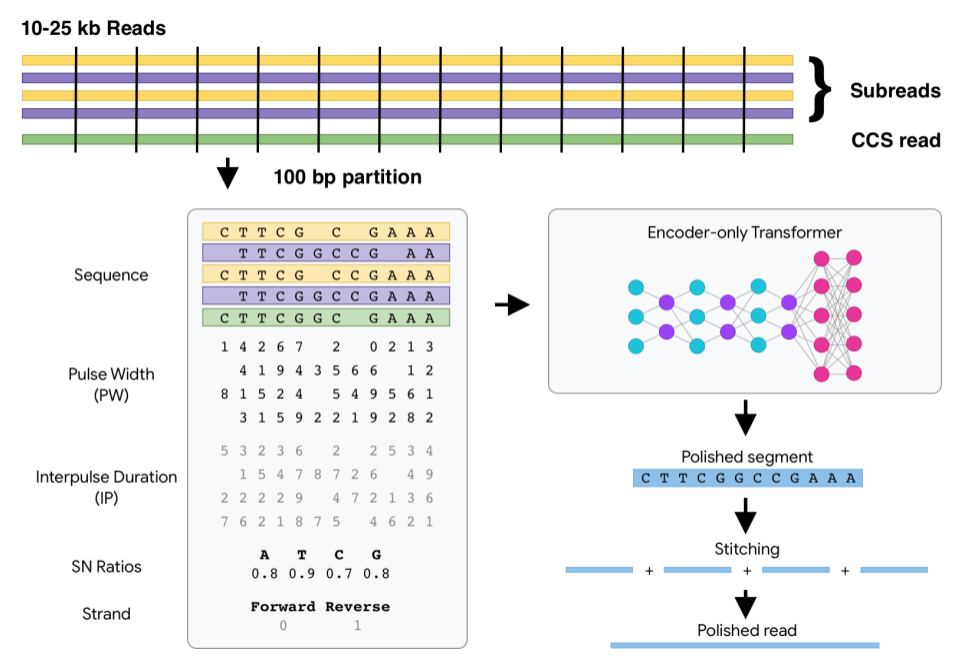
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
If you're on a GPU machine:
pip install deepconsensus[gpu]==0.2.0
# To make sure the `deepconsensus` CLI works, set the PATH:
export PATH="/home/${USER}/.local/bin:${PATH}"If you're on a CPU machine:
pip install deepconsensus[cpu]==0.2.0
# To make sure the `deepconsensus` CLI works, set the PATH:
export PATH="/home/${USER}/.local/bin:${PATH}"For GPU:
sudo docker pull google/deepconsensus:0.2.0-gpuFor CPU:
sudo docker pull google/deepconsensus:0.2.0git clone https://github.com/google/deepconsensus.git
cd deepconsensus
source install.shIf you have GPU, run source install-gpu.sh instead. Currently the only
difference is that the GPU version installs tensorflow-gpu instead of
intel-tensorflow.
(Optional) After source install.sh, if you want to run all unit tests, you can
do:
./run_all_tests.shSee the quick start.
After a PacBio sequencing run, DeepConsensus is meant to be run on the subreads to create new corrected reads in FASTQ format that can take the place of the CCS reads for downstream analyses.
See the quick start for an example of inputs and outputs.
If you are using DeepConsensus in your work, please cite:
DeepConsensus: Gap-Aware Sequence Transformers for Sequence Correction
This is not an official Google product.
NOTE: the content of this research code repository (i) is not intended to be a medical device; and (ii) is not intended for clinical use of any kind, including but not limited to diagnosis or prognosis.