Python implementation of SINGLE algorithm. Full details can be found [here] (http://www.sciencedirect.com/science/article/pii/S1053811914006168) or alternatively [here] (http://mirrors.softliste.de/cran/web/packages/SINGLE/vignettes/vignette.pdf)
The SINGLE class has the following methods (see example below):
fit_radius: this is used to estimate the radius of the Gaussian kernel, h.tune_params: this function is used to estimate sparsity, l1, and temporal homogeneity, l2, parametersfit: Once parameters h, l1 and l2 have been estimated this function can be used to estimate an array of sparse precision matrices using the SINGLE algorithm.plot: used to plot resulting partial correlations
After downloading the ZIP file:
sudo python setup.py install
We provide simulated data in the Sample Data folder.
import numpy, pandas
from pySINGLE.SINGLE import SINGLE
# read in data:
data = numpy.array(pandas.read_csv('pySINGLE/Sample Data/biggerdata.csv'))
# establish instance of SINGLE class:
S = SINGLE(data=data)
# estimate width of Gaussian kernel, h:
S.fit_radius(h_vals=[10,12.5, 15], samples=None)
# estimate sparsity and temporal homogeneity parameters:
S.tune_params()
# finally, we can fit the model according to the estimated parameters:
S.fit()
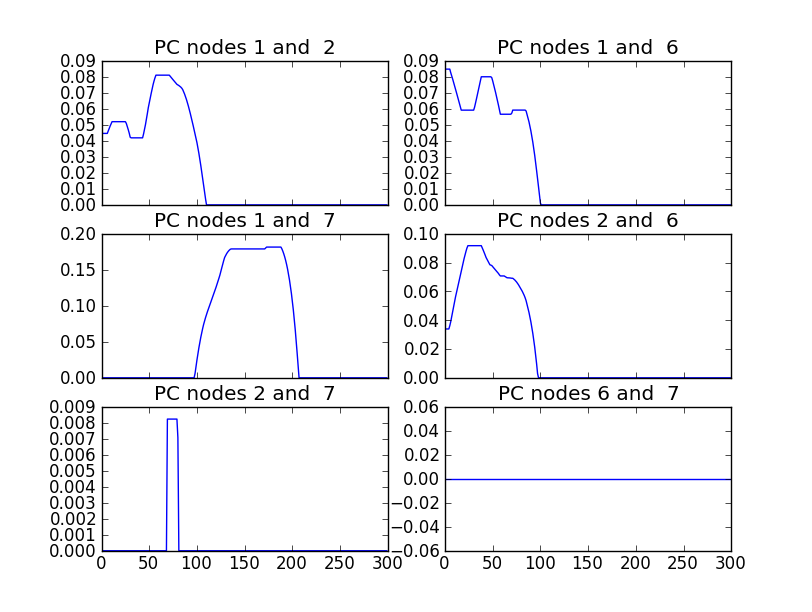
# and plot partial correlations between nodes 1,2,6 and 7 (image below)
S.plot(index=[1,2,6,7], ncol_=2)