In many situations, the user interaction is superfluous and the desired goal is
to simply display a secondary structure on a web page. This is a common
scenario in, for example, servers that predict a secondary structure. The
output, a dot-bracket string can simply be added to a FornaContainer object
to display.
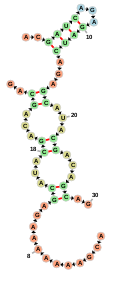
Below is an example of a simple web page which uses a FornaContainer to show
a simple RNA molecule:
The code for creating this web page is rather straightforward. After importing
some necessary javascript files, we create a container using new FornaContainer("#rna_ss", {'animation': false}), passing in #rna_ss as the
id of the div which will hold the container and then populate it with a
structure and sequence using container.addRNA:
<!DOCTYPE html>
<meta charset="utf-8">
This is an RNA container.
<div id='rna_ss'> </div>
This after the RNA container.
<link rel='stylesheet' type='text/css' href='fornac.css' />
<script type='text/javascript' src='fornac.js'></script>
<script type='text/javascript'>
var container = new FornaContainer("#rna_ss", {'animation': false});
var options = {'structure': '((..((....)).(((....))).))',
'sequence': 'CGCUUCAUAUAAUCCUAAUGACCUAU'
};
container.addRNA(options.structure, options);
</script>Display two cofolded sequences using the format of RNAcofold:
var container = new fornac.FornaContainer("#cofold_ss",
{'animation': false, 'zoomable': true, 'initialSize':[500,300]});
var options = {'structure': '..((((...))))...((...((...((..&............))...))...))..',
'sequence': 'ACGAUCAGAGAUCAGAGCAUACGACAGCAG&ACGAAAAAAAGAGCAUACGACAGCAG'
};
container.addRNA(options.structure, options);
container.setSize(); fornac.getStructuresDotBracket()
Returns a dot-bracket representation of the visible structure. This is useful
if integrating this component with structure editing functionality. Returns an
array of length 2 (e.g. ['CCCCAAAAGGGG', '((((....))))']).
The FornaContainer supports a number of options to allow users to customize how the RNA will be presented.
Indicate whether the force-directed layout will be applied to the displayed molecule. Enabling this option allows users to change the layout of the molecule by selecting and dragging the individual nucleotide nodes
Allow users to zoom in and pan the display. If this is enabled then pressing the 'c' key on the keyboard will center the view.
This only makes sense in connection with the animation argument. If it's
true, the external loops will be arranged in a nice circle. If false, they will
be allowed to flop around as the force layout dictates:
Change how often nucleotide numbers are labelled with their number.
fornac.getStructuresDotBracket()
Yields
["AAAACCCC&UUUUGGGG", ".(....).&.(....)."]
Each RNA molecule is represented as a JSON file which encodes all of the
information necessary to display it. The example shows a trivial and slightly
modified example. nodeType can be either nucleotide or label or middle,
the latter of which is used only as a placeholder for maintaining an aesthetically
pleasing layout.
The links can be any of basepair (representing a basepair between two
nucleotides), backbone (backbone bond between adjacent nodes), pseudoknot
(pseudoknot, extracted from the specified structure using maximum matching
algorithm), extra (extra links specified by the user), label_link (links
between nucleotides and their nucleotide number labels), fake (invisible
links for maintaining the layout), and fake_fake (invisible links for
maintaining the layout).
This structure is initially created in rnagraph.js starting from a sequence
and dotbracket string.
{
"nodes":
[ {
"name": "A",
"num": 1,
"radius": 5,
"rna": null,
"nodeType": "nucleotide",
"structName": "empty",
"elemType": "e",
"uid": "44edb966-aca9-4058-a6bc-784a34959329",
"linked": false,
"prevNode": null,
"nextNode": null,
"x": 100,
"px": 100,
"y": 100,
"py": 100
},
...
],
"links":
[ {
"source": null,
"target": null,
"linkType": "basepair",
"value": 1,
"uid": "6664a569-5af1-4d86-8ada-d1c00da72a899f87a224-52a0-4ede-a29c-04fddc09e4c4"
},
...
]
}
First:
npm install
To debug and to show examples:
npm run dev
To build minified distribution:
npm run build
The output will be placed in the dist directory. To use fornac in a web page, simply include dist/fornac.js and the css style dist/fornac.css in your web page.
Thanks to Benedikt Rauscher for the Javascript version of the NAView layout and to the creators of VARNA for the java implementation that it's based on.