-
Notifications
You must be signed in to change notification settings - Fork 6
Binning
BinaRena, as the name suggests, is dedicated to metagenomic binning. It provides various features to facilitate this process.
One may create a binning plan from de novo or load an existing one from a categorical field. Each category represents a bin. The name, number of contigs (#), total length of contigs (kb) and the relative abundance (calculated as length x coverage, then normalized against the entire assembly) are displayed.
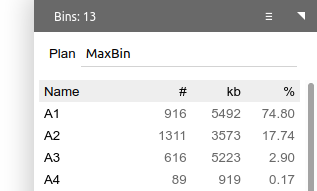
When a binning plan is modified (added/removed bins, added/removed contigs to/from bins, renamed bins, entered a new plan name, etc.), a Save button will show up. Click it to save the binning plan as a categorical field.
Alternatively, click the ⇥ button in the toolbar to export the binning plan to a text file. The format will be a mapping of bin names to member contig IDs.
The table below the binning plan name displays a list of bins in the plan. Click a bin to select it. The contigs in this bin will be selected at the same time and their properties will be summarized.
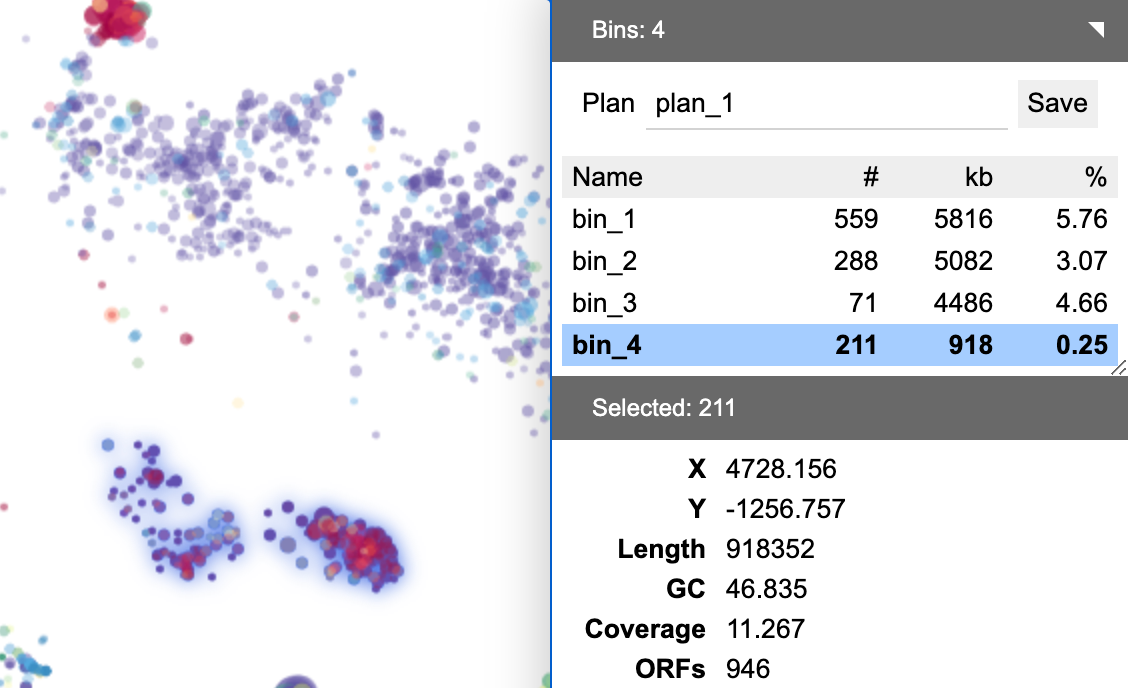
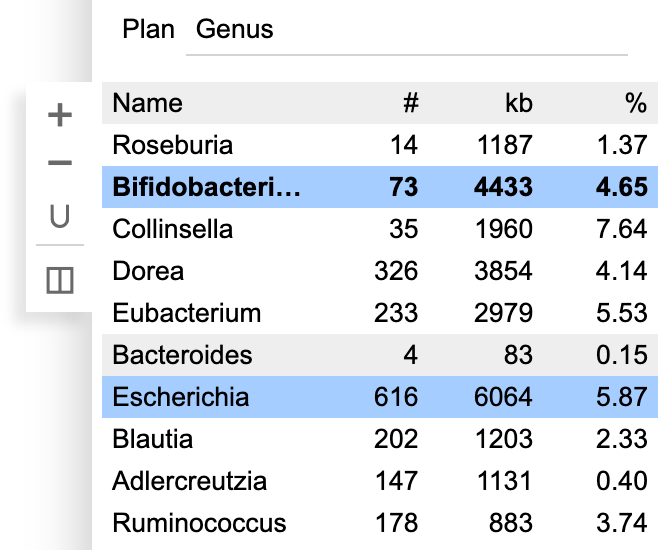
Click the bin again to edit its name. Press Enter when done. Hold Shift and click to select multiple bins.
The toolbar next to the bin table provides a few utilities. Click + to create an empty new bin. Click − to delete the selected bin(s), Click ∪ to merge multiple bins.
Press Space to create a new bin from selected contigs. If no binning plan is currently loaded, the program will create a new plan. This is perhaps the easiest way to start de novo binning using the program.
Press . to add selected contigs to the select bin. Press , to remove selected contigs from the selected bin. Press / to update (replace) the selected bin with selected contigs.
These functions can also be found in the toolbar next to the summary table.
See also how to export binning results.
Contact: Dr. Qiyun Zhu (qiyunzhu@gmail.com).