-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
RGB to HED conversion after version 0.17 #5164
Comments
|
Hi @nicolebussola thanks for reporting this. It's true that the doc example https://scikit-image.org/docs/dev/auto_examples/color_exposure/plot_ihc_color_separation.html does not look great. @alexdesiqueira since you worked on this PR could you please comment? |
|
Hi @nicolebussola, thank you for opening this issue. Yeah @emmanuelle, the doc example doesn't "feel right" anymore.
That said, how does that sound, Nicole? Is there something I'm missing, or would you like to add something else? |
|
In Current code for rgb = _prepare_colorarray(rgb, force_copy=True)
np.maximum(rgb, 1E-6, out=rgb) # avoiding log artifacts
log_adjust = np.log(1E-6) # used to compensate the sum above
stains = (np.log(rgb) / log_adjust) @ conv_matrixIn the last line, np.minimum(stains, 0, out= stains)should fix the function. If we create the input artificially by combining positive values and using the colors as assumed in Yes, this clipping makes the round-trip not idempotent. But if you start with a synthetic image, the clip will do nothing and hence the tests should continue to work. On the other hand, ihc_hed = rgb2hed(ihc_rgb)
reconstructed = hed2rgb(ihc_hed)shows in |
|
Also, the example at https://scikit-image.org/docs/dev/auto_examples/color_exposure/plot_ihc_color_separation.html has a bug: cmap_eosin = LinearSegmentedColormap.from_list('mycmap', ['darkviolet', 'white'])The two colors are reversed, this should be: cmap_eosin = LinearSegmentedColormap.from_list('mycmap', ['white', 'darkviolet'])Nobody noticed this before because the image has no Eosin, and so this channel has only residue (= negative values). Reversing the colormap actually looks good in this case, but the image should be all zeros (white), which we do see after clipping negative values. Finally, the DAB channel (brown) looks rather dim. This is because there is one pixel that happens to be really dense (maybe noise?) and the image is scaled to that. But this is just a display issue. If you want to avoid this, you can do like in the ImageJ plugin, where they don't use 1e-6 but 1/255 in the initial clip. This causes the darkest stain to be less dark. This is a question about what you prefer: better display or better data. |
|
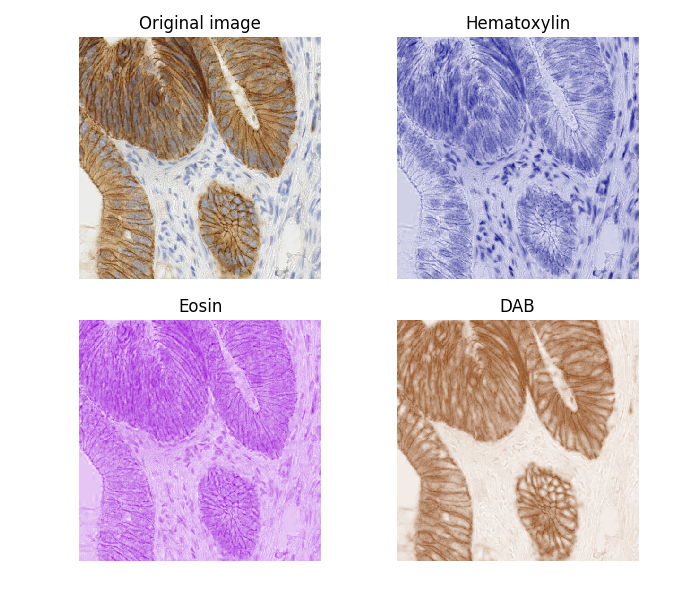
OK, one last comment from me and then I'll shut up. :) I'd change the demo as follows for proper display of the unmixed stains. What we're doing here is creating an image in "hed color space" that has only one stain, then using import matplotlib.pyplot as plt
import numpy as np
from skimage import data
from skimage.color import rgb2hed, hed2rgb
ihc_rgb = data.immunohistochemistry()
ihc_hed = rgb2hed(ihc_rgb)
ihc_hed[ihc_hed<0] = 0 # TODO: remove this after fixing `rgb2hed()`
null = np.zeros_like(ihc_hed[:,:,0])
ihc_h = hed2rgb(np.stack((ihc_hed[:,:,0], null, null), axis=-1))
ihc_e = hed2rgb(np.stack((null, ihc_hed[:,:,1], null), axis=-1))
ihc_d = hed2rgb(np.stack((null, null, ihc_hed[:,:,2]), axis=-1))
fig, axes = plt.subplots(2, 2, figsize=(7, 6), sharex=True, sharey=True)
ax = axes.ravel()
ax[0].imshow(ihc_rgb)
ax[0].set_title("Original image")
ax[1].imshow(ihc_h)
ax[1].set_title("Hematoxylin")
ax[2].imshow(ihc_e)
ax[2].set_title("Eosin")
ax[3].imshow(ihc_d)
ax[3].set_title("DAB")
for a in ax.ravel():
a.axis('off')
fig.tight_layout()
plt.show()It is still fine to use |
|
Thanks for sorting this out @crisluengo! This is looking much better and the fix is pretty straightforward. Let us know if you want to make a PR, otherwise we can take care of it. |
|
Hi @nicolebussola, please let us know if the current code solves these issues. Thank you again @crisluengo! |
* resize using scipy.ndimage.zoom * Clip negative values in output of RGB to HED conversion. Fixes #5164 * Fixed stain separation tests. - Round trips start with stains, such that the roundtrip does not require negative values. - Using a blue+red+orange stain combination, the test previously used a 2-stain combination that made it impossible to do a correct roundtrip. * Improve IHC stain separation example. * BENCH: add benchmarks for resize and rescale * move mode conversion code to _shared.utils.py use grid-constant and grid-wrap for SciPy >= 1.6.0 closes gh-5064 * separate grid mode conversion utility * Update doc/examples/color_exposure/plot_ihc_color_separation.py Improvement to the IHC example. Co-authored-by: Riadh Fezzani <rfezzani@gmail.com> * add TODO for removing legacy code paths * avoid pep8 warning in benchmark_interpolation.py * keep same ValueError on unrecognized modes * test_rank.py: fetch reference data once into a dictionary of arrays (#5175) This should avoid stochastic test failures that have been occuring on GitHub Actions * Add normalized mutual information metric (#5158) * Add normalized mutual information metric * Add tests for NMI * Add NMI to metrics init * Fix error message for incorrect target shape * Add 3D NMI test * properly add nmi to metrics init * _true, _test -> 0, 1 * ravel -> reshape(-1) * Apply suggestions from @mkcor's code review Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org> * Fix warning and exception messages Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org> * segment Other section of TODO by date based on NEP-29 24 month window for dropping upstream package version * Fix bug in optical flow code and examples mode 'nearest' is a scipy.ndimage mode, but warp only accepts the numpy.pad equivalent of 'edge' * fix additional cases of warp with 'nearest'->'edge' * DOC: Fixup to linspace in example (#5181) Right now the data is not sampled every half degree. Also, -pi/2 to pi/2. * Minor fixups to Hough line transform code and examples (#5182) * Correction to linspace call in hough_line default `theta = np.pi / 2 - np.arange(180) / 180.0 * np.pi` done correctly below. * BUG: Fixup to image orientation in example * DOC: Tiny typo * Changed range to conform to documentation The original was [pi/2, -pi/2), not [-pi/2, pi/2). Updated wording in docs. * Fixed x-bounds of hough image in example * Fixed long line * Fixes to tests Number of lines in checkerboard, longer line in peak ordering assertion, updated angle * Accidentally modified the wrong value. * Updated example to include three lines As requested in review * Fixed off-by one error in pixel bins in hough line transform (#5183) * Added 1/2 pixel bounds to extent of displayed images (#5184) * fast, non-Cython implementation for correlate_sparse (#5171) * Replace correlate_sparse Cython version with a simple slicing-based variant in _mean_std This slicing variant has two benefits: 1.) profiled 30-40% faster on 2d and 3d cases I tested with 2.) can immediately be used by other array backends such as CuPy * remove intermediate padded_sq variable to reduce memory footprint * Update skimage/filters/_sparse.py remove duplicate declarations Co-authored-by: Riadh Fezzani <rfezzani@gmail.com> * move _to_np_mode and _to_ndimage_mode to _shared.utils.py * add comment about corner_index needing to be present * raise RuntimeError on missing corner index * update code style based on reviewer comments Co-authored-by: Riadh Fezzani <rfezzani@gmail.com> * Add release step on github to RELEASE.txt (See #5185) (#5187) Co-authored-by: Juan Nunez-Iglesias <juan.nunez-iglesias@monash.edu> * Prevent integer overflow in EllipseModel (#5179) * Change dtype of CircleModel and EllipseModel to float, to avoid integer overflows * remove failing test and add code review notes * Apply suggestions from code review Co-authored-by: Stefan van der Walt <sjvdwalt@gmail.com> * Remove unused pytest import Co-authored-by: Stefan van der Walt <sjvdwalt@gmail.com> Co-authored-by: Stefan van der Walt <sjvdwalt@gmail.com> Co-authored-by: Juan Nunez-Iglesias <juan.nunez-iglesias@monash.edu> * Add saturation parameter to color.label2rgb (#5156) * Remove reference to opencv in threshold_local documentation (#5191) Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org> Co-authored-by: Riadh <r.fezzani@vitadx.com> * resize using scipy.ndimage.zoom * BENCH: add benchmarks for resize and rescale * move mode conversion code to _shared.utils.py use grid-constant and grid-wrap for SciPy >= 1.6.0 closes gh-5064 * separate grid mode conversion utility * add TODO for removing legacy code paths * avoid pep8 warning in benchmark_interpolation.py * keep same ValueError on unrecognized modes * segment Other section of TODO by date based on NEP-29 24 month window for dropping upstream package version * Fix bug in optical flow code and examples mode 'nearest' is a scipy.ndimage mode, but warp only accepts the numpy.pad equivalent of 'edge' * fix additional cases of warp with 'nearest'->'edge' * Apply suggestions from code review Co-authored-by: Riadh Fezzani <rfezzani@gmail.com> Co-authored-by: Cris Luengo <cris.l.luengo@gmail.com> Co-authored-by: Cris Luengo <crisluengo@users.noreply.github.com> Co-authored-by: Riadh Fezzani <rfezzani@gmail.com> Co-authored-by: Alexandre de Siqueira <alex.desiqueira@igdore.org> Co-authored-by: Juan Nunez-Iglesias <juan.nunez-iglesias@monash.edu> Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org> Co-authored-by: Joseph Fox-Rabinovitz <madphysicist@users.noreply.github.com> Co-authored-by: François Boulogne <fboulogne@sciunto.org> Co-authored-by: Mark Boer <m.h.boer.2@gmail.com> Co-authored-by: Stefan van der Walt <sjvdwalt@gmail.com> Co-authored-by: Carlos Andrés Álvarez Restrepo <charlie_cha@outlook.com> Co-authored-by: Riadh <r.fezzani@vitadx.com>


Description
After upgrading our project environment to version 0.18.x we have noticed that the RGB to HED conversion behaves unexpectedly according to the biological meaning of the HED color space. We have done some research and carefully read the PR changing the stain deconvolution method, however the HED conversion in version 0.17.x seemed to be more appropriate; in particular the H channel highlights the nuclei while the DAB channel highlights the brown stain:
version 0.17.x
We are not able to find the same biological meaning in version 0.18.x:
version 0.18.x

Regards,
Nicole, Alessia, Ernesto (histolab team)
The text was updated successfully, but these errors were encountered: