New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[MRG+2] Neighborhood Components Analysis #10058
Conversation
Here is a snippet that shows it (on my machine which has 7.7GB of memory): from sklearn.datasets import load_digits
from metric_learn import NCA
from sklearn.neighbors import NeighborhoodComponentsAnalysis
from sklearn.utils.testing import assert_raises
digits = load_digits()
X, y = digits.data, digits.target
nca_ml = NCA()
assert_raises(MemoryError, nca_ml.fit, X, y)
nca_sk = NeighborhoodComponentsAnalysis()
nca_sk.fit(X, y) # does not raise any error |
Indeed, they do not add a significative speedup but have a high memory cost.
sklearn/neighbors/nca.py
Outdated
| transformation = transformation.reshape(-1, X.shape[1]) | ||
| loss = 0 | ||
| gradient = np.zeros(transformation.shape) | ||
| X_embedded = transformation.dot(X.T).T |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
With np.dot(X, transformation.T) you save a transpose.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
True, I will change it.
|
Hi @wdevazelhes , since the code is very similar to LMNN, I felt free to add just a couple of comments, by no means a full review. @jnothman , I hope I didn't break any contribution guidelines.. |
sklearn/neighbors/nca.py
Outdated
| diff_embedded[~ci, :])) | ||
| p_i = np.sum(p_i_j) # probability of x_i to be correctly | ||
| # classified | ||
| gradient += 2 * (p_i * (sum_ci.T + sum_not_ci.T) - sum_ci.T) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You could just add the matrices without transposing, and transpose the gradient once before returning the unraveled gradient.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I remembered I had seen a more efficient computation of the gradient (see last equation in slide 12 here: http://bengio.abracadoudou.com/lce/slides/roweis.pdf ).
This would amount to:
p_i_j = exp_dist_embedded[ci]
p_i_k = exp_dist_embedded[~ci]
diffs = X[i, :] - X
diff_ci = diffs[ci, :]
diff_not_ci = diffs[~ci, :]
sum_ci = diff_ci.T.dot(p_i_j[:, np.newaxis] * diff_ci)
sum_not_ci = diff_not_ci.T.dot(p_i_k[:, np.newaxis] * diff_not_ci)
p_i = np.sum(p_i_j)
gradient += p_i * sum_not_ci + (1 - p_i) * sum_ci
And multiplying by the transformation after the for-loop:
gradient = 2*np.dot(transformation, gradient)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You could just add the matrices without transposing, and transpose the gradient once before returning the unraveled gradient.
True, I will change it.
I remembered I had seen a more efficient computation of the gradient (see last equation in slide 12 here: http://bengio.abracadoudou.com/lce/slides/roweis.pdf ).
This would amount to:
[...]
And multiplying by the transformation after the for-loop:
gradient = 2*np.dot(transformation, gradient)
Since I already computed embedded differences for the softmax before wouldn't it be more efficient to reuse it ?
I agree that the expression with (1 - p_i) is clearer though.
Codecov Report
@@ Coverage Diff @@
## master #10058 +/- ##
==========================================
+ Coverage 96.19% 96.21% +0.01%
==========================================
Files 336 338 +2
Lines 62740 63025 +285
==========================================
+ Hits 60354 60638 +284
- Misses 2386 2387 +1
Continue to review full report at Codecov.
|
|
Thanks for your comments @johny-c ! I have modified the code according to them. |
|
I also added an example: plot_nca_dim_reduction.py (in commit 12cf3a9) |
|
I benchmarked this PR implementation of NCA against metric-learn one: I plotted the training curves (objective function vs time), for the same initialisation (identity), on At some points, metric-learn NCA training is interrupted prematurely: this is due to numerical instabilities, and this warning is thrown: |
| # Plot the embedding and show the evaluation score | ||
| plt.scatter(X_embedded[:, 0], X_embedded[:, 1], c=y) | ||
| plt.title("{}, KNN (k={})".format(name, n_neighbors)) | ||
| plt.text(0.9, 0.1, '{:.2f}'.format(acc_knn), size=15, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The accuracy is a bit misplaced when running the example on my laptop. It is probably probably easier to put "Test accuracy = x" in the title (after a line break)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done
sklearn/neighbors/nca.py
Outdated
| import time | ||
| from scipy.misc import logsumexp | ||
| from scipy.optimize import minimize | ||
| from sklearn.preprocessing import OneHotEncoder |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
use relative import
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done
|
|
||
| pca: | ||
| ``n_features_out`` many principal components of the inputs passed | ||
| to :meth:`fit` will be used to initialize the transformation. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
all params are not indented the same way
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is because pca, identity, random and numpy array are not arguments but they are possible choices for argument init. I took the syntax from LMNN. Should I write it in another way ?
sklearn/neighbors/nca.py
Outdated
| ---------- | ||
| transformation : array, shape (n_features_out, n_features) | ||
| The linear transformation on which to compute loss and evaluate | ||
| gradient |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
insert empty line
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done
sklearn/neighbors/nca.py
Outdated
| X_embedded = np.dot(X, transformation.T) | ||
|
|
||
| # for every sample x_i, compute its contribution to loss and gradient | ||
| for i in range(X.shape[0]): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
looping over samples seems a problem. Is it possible to vectorize?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
it should be possible but one should then try to avoid an O(n_samples*n_samples*n_features) memory complexity
|
maybe use minibatches?
|
sklearn/neighbors/nca.py
Outdated
|
|
||
| Parameters | ||
| ---------- | ||
| transformation : array, shape(n_components, n_features) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
is it raveled here or not?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Absolutely, it should be raveled, good catch, I'll change it
| @@ -0,0 +1,511 @@ | |||
| import pytest | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I would add here a header with license and authors on top of the file like we do in other places
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
That's right it's missing, but did you mean to say in the main file nca.py ? I looked in two or three tests code and it didn't seem they had an author section/licence
|
ok did one (last?) round. more tomorrow! |
|
tell me when ready
… |
| @@ -3,7 +3,9 @@ | |||
| Neighborhood Component Analysis | |||
| """ | |||
|
|
|||
| # License: BSD 3 Clause | |||
| # Authors: William de Vazelhes <wdevazelhes@gmail.com> | |||
| # John Chiotellis <ioannis.chiotellis@in.tum.de> | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@johny-c I put the email I found on your webpage, but feel free to put another one here ;)
|
There just remains to settle on #10058 (comment), and check on #10058 (comment) and maybe wait for @johny-c to put his email (otherwise he could do it after) ? Otherwise I addressed all your comments, so, it's ready @agramfort @GaelVaroquaux ! |
| """ | ||
|
|
||
| # Authors: William de Vazelhes <wdevazelhes@gmail.com> | ||
| # John Chiotellis <ioannis.chiotellis@in.tum.de> |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@johny-c same remark here too
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM too :-)
|
LGTM. +1 for merge once the CI has run. |
|
Let's do it |
|
Congratulations @wdevazelhes!!!! This has been a long time coming... |
|
That's great ! Thanks a lot for your reviews and comments @jnothman @agramfort @GaelVaroquaux @bellet, and congrats to @johny-c too ! I'm excited to work on improvements later on. Also to transpose the changes of this PR to LMNN so that it can be merged, or maybe we'll see first how NCA goes for a bit of time and then see what to do for LMNN |
|
Congrats @wdevazelhes ! Indeed, core devs should decide whether they also want to include LMNN (#8602). I think it would be good as LMNN is also a popular method and there is quite a lot of code that could be shared with NCA. |
|
Hi all! @wdevazelhes, congrats and thanks for including me as an author. Regarding LMNN, as far as I remember, what is missing is some testing . I will have a couple of free weeks in March, so I could have another look at it. |
This reverts commit 5e8a4ea.
This reverts commit 5e8a4ea.
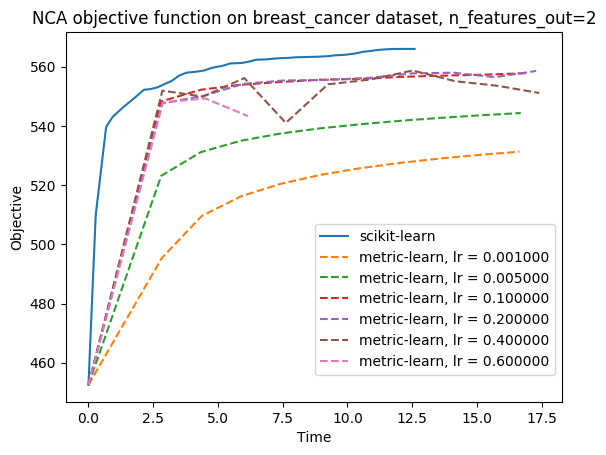
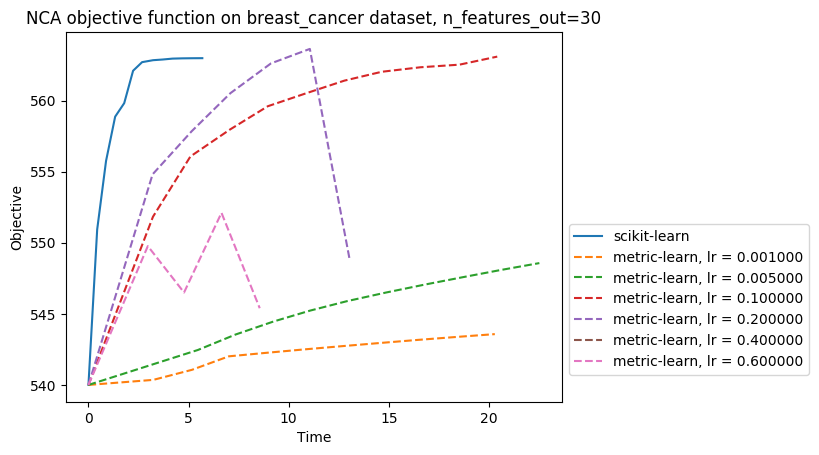
Hi, this PR is an implementation of the Neighborhood Components Analysis algorithm (NCA), a popular supervised distance metric learning algorithm. As LMNN (cf PR #8602) this algorithm takes as input a labeled dataset, instead of similar/dissimilar pairs like it is the case for most metric learning algorithms, and learns a linear transformation of the space. However, NCA and LMNN have different objective functions: NCA tries to maximise the probability of every sample to be correctly classified based on a stochastic nearest neighbors rule, and therefore does not need to fix in advance a set of target neighbors.
There have been several attempts to implement NCA (2 PRs: #5276 (closed) and #4789 (not closed)). I created a fresh PR for sake of clarity. Indeed, this code is intended to be as similar to LMNN as possible, which should allow the factorisation of some points of code which are the same in both algorithms.
At the time of writing, this algorithm uses scipy's L-BFGS-B solver to solve the optimisation problem, like LMNN. It has the big advantage of avoiding to tune a learning rate parameter.
I benchmarked this implementation to the package metric-learn's one (https://github.com/all-umass/metric-learn): the one in this PR has the advantage of being scalable to large datasets (indeed, metric-learn's NCA throws
Memory Errorfor too big datasets like faces or digits), for no significative loss in performance for small datasets.The remaining tasks are the following:
instance metric learn's one) (coming soon)
What is more, some improvements could also be made in a second time:
datasets (probably in another PR ?)
Feedback is welcome !