-
Notifications
You must be signed in to change notification settings - Fork 2
Home
##Non-negative Matrix Factorization Non-negative matrix factorization is the process of decomposing a two dimensional dense matrix, V, into two matrices of lower dimensionality, W and H, with all three matrices having no-negative values. W and H share a dimension, k, which represents the number of components (or features) of V which are being represented in the decomposed matrices. NMF is not a specific algorithm but instead a family of algorithms with a variety of implementations. These algorithms have applications in fields such as computer vision, audio processing, recommendation systems and a variety of other domains.
##The problem
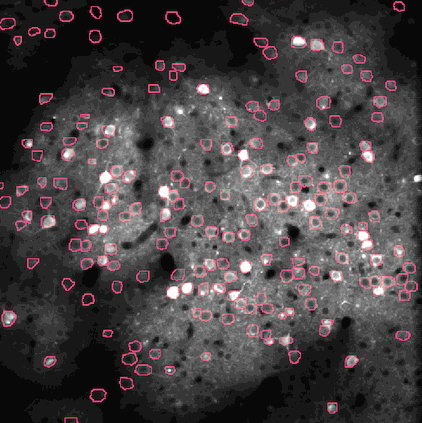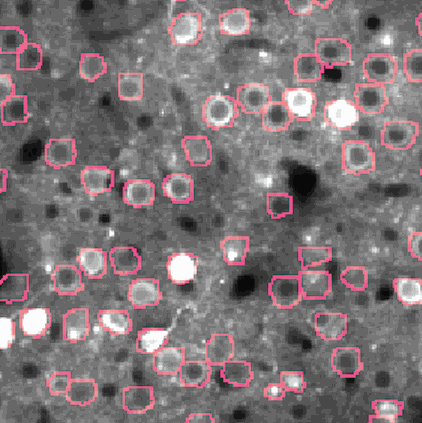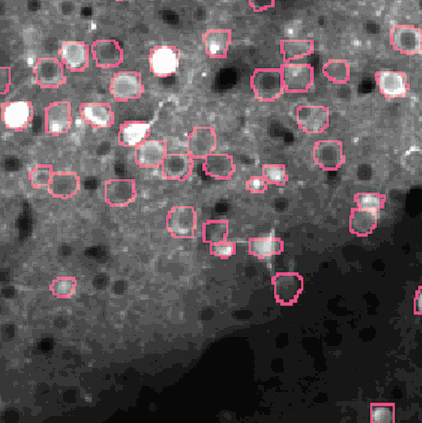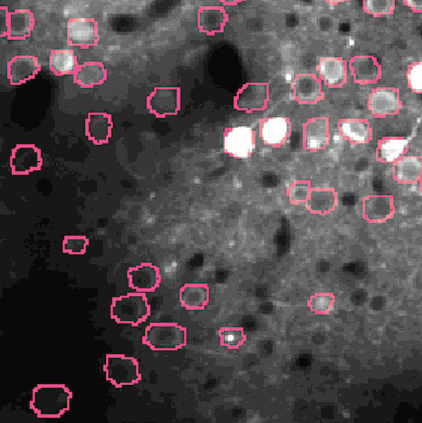
Calcium imaging is a technique for identifying neurons. As calcium is inject the neurons activate, resulting in the neurons flashing white in the generated images. The issue is then identifying the individual neurons in this densely packed, complex system of cell. This problem set is known as neurofinder. Given a video (or time series of still images) of a section of brain, identify and output the number, center, and border of neurons.
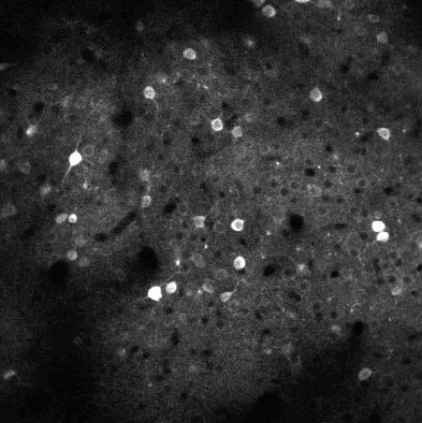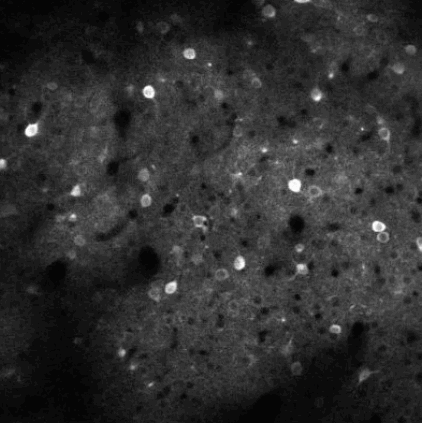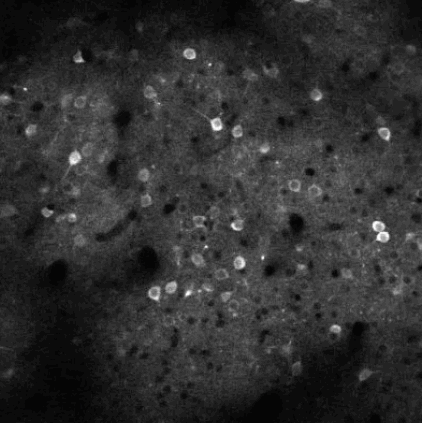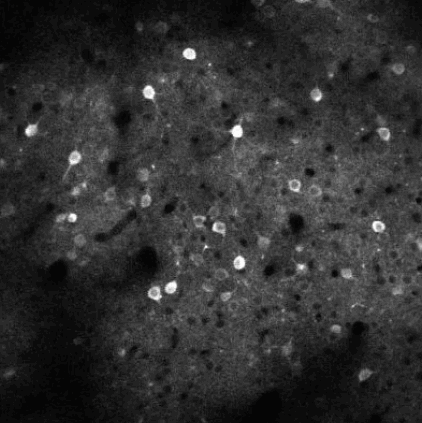
##Our approach We used a library know as Thunder. This is library is a tool set designed specifically for this domain of problems and is an excellent option for our processing of the neurofinder data set. Thunder has a package containing a variety of matrix factorization algorithms, which we used for our non-negative matrix factorization of our data. Using Thunder along side Spark, which thunder supports, we were able to distribute the NMF process among each from of the neuron data in order to receive our feature matrices from the training data, then used the resulting matrices for identifying neurons in the testing data.