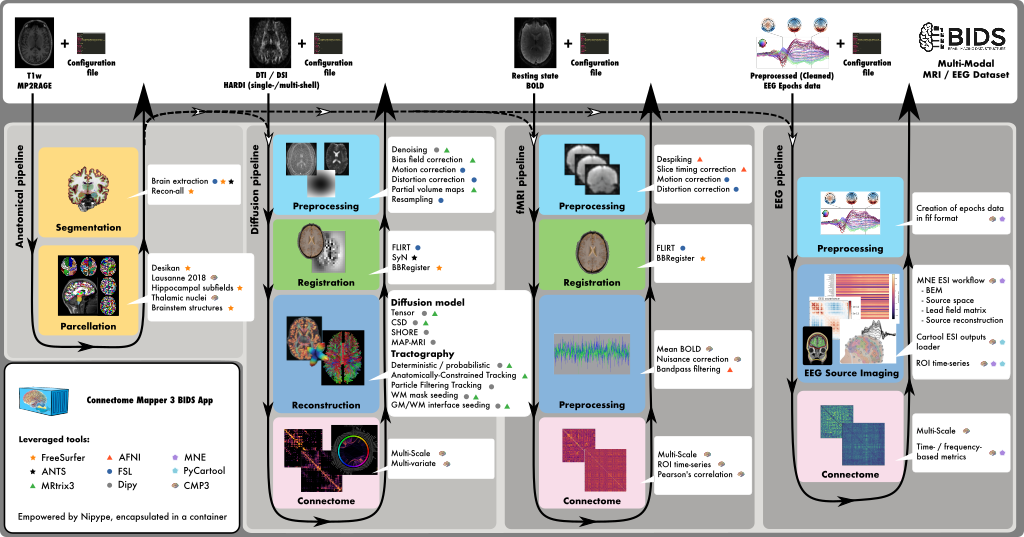
This neuroimaging processing pipeline software is developed by the Connectomics Lab at the University Hospital of Lausanne (CHUV) for use within the SNF Sinergia Project 170873, as well as for open-source software distribution.
Connectome Mapper 3 is an open-source Python3 image processing pipeline software that implements full anatomical, diffusion and resting-state MRI processing pipelines, from raw Diffusion / T1 / T2 / BOLD data to multi-resolution connection matrices.
Connectome Mapper 3 pipelines use a combination of tools from well-known software packages, including FSL, FreeSurfer, ANTs, MRtrix3, Dipy and AFNI, orchestrated by the Nipype dataflow library. These pipelines were designed to provide the best software implementation for each state of processing, and will be updated as newer and better neuroimaging software become available.
This tool allows you to easily do the following:
- Take T1 / Diffusion / resting-state MRI data from raw to multi-resolution connection matrices.
- Implement tools from different software packages.
- Achieve optimal data processing quality by using the best tools available
- Automate and parallelize processing steps, providing a significant speed-up from typical linear, manual processing.
Reproducibility and replicatibility is achieved through the distribution of a BIDSApp, a software container image which takes BIDS datasets as inputs and which provides a frozen environment where versions of all external softwares and libraries are fixed.
- Documentation: https://connectome-mapper-3.readthedocs.io
- Mailing list: https://groups.google.com/forum/#!forum/cmtk-users
- Source: https://github.com/connectomicslab/connectomemapper3
- Bug reports: https://github.com/connectomicslab/connectomemapper3/issues
This BIDS App has the following command line arguments:
$ docker run -it sebastientourbier/connectomemapper-bidsapp -h
usage: run.py [-h]
[--participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]]
[--session_label SESSION_LABEL [SESSION_LABEL ...]]
[--anat_pipeline_config ANAT_PIPELINE_CONFIG]
[--dwi_pipeline_config DWI_PIPELINE_CONFIG]
[--func_pipeline_config FUNC_PIPELINE_CONFIG]
[--number_of_threads NUMBER_OF_THREADS]
[--number_of_participants_processed_in_parallel NUMBER_OF_PARTICIPANTS_PROCESSED_IN_PARALLEL]
[--mrtrix_random_seed MRTRIX_RANDOM_SEED]
[--ants_random_seed ANTS_RANDOM_SEED]
[--ants_number_of_threads ANTS_NUMBER_OF_THREADS]
[--fs_license FS_LICENSE] [--coverage] [--notrack] [-v]
bids_dir output_dir {participant,group}
Entrypoint script of the BIDS-App Connectome Mapper version v3.0.0-RC3
positional arguments:
bids_dir The directory with the input dataset formatted
according to the BIDS standard.
output_dir The directory where the output files should be stored.
If you are running group level analysis this folder
should be prepopulated with the results of the
participant level analysis.
{participant,group} Level of the analysis that will be performed. Multiple
participant level analyses can be run independently
(in parallel) using the same output_dir.
optional arguments:
-h, --help show this help message and exit
--participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]
The label(s) of the participant(s) that should be
analyzed. The label corresponds to
sub-<participant_label> from the BIDS spec (so it does
not include "sub-"). If this parameter is not provided
all subjects should be analyzed. Multiple participants
can be specified with a space separated list.
--session_label SESSION_LABEL [SESSION_LABEL ...]
The label(s) of the session that should be analyzed.
The label corresponds to ses-<session_label> from the
BIDS spec (so it does not include "ses-"). If this
parameter is not provided all sessions should be
analyzed. Multiple sessions can be specified with a
space separated list.
--anat_pipeline_config ANAT_PIPELINE_CONFIG
Configuration .txt file for processing stages of the
anatomical MRI processing pipeline
--dwi_pipeline_config DWI_PIPELINE_CONFIG
Configuration .txt file for processing stages of the
diffusion MRI processing pipeline
--func_pipeline_config FUNC_PIPELINE_CONFIG
Configuration .txt file for processing stages of the
fMRI processing pipeline
--number_of_threads NUMBER_OF_THREADS
The number of OpenMP threads used for multi-threading
by Freesurfer (Set to [Number of available CPUs -1] by default).
--number_of_participants_processed_in_parallel NUMBER_OF_PARTICIPANTS_PROCESSED_IN_PARALLEL
The number of subjects to be processed in parallel
(One by default).
--mrtrix_random_seed MRTRIX_RANDOM_SEED
Fix MRtrix3 random number generator seed to the
specified value
--ants_random_seed ANTS_RANDOM_SEED
Fix ANTS random number generator seed to the specified
value
--ants_number_of_threads ANTS_NUMBER_OF_THREADS
Fix number of threads in ANTs. If not specified ANTs
will use the same number as the number of OpenMP
threads (see `----number_of_threads` option flag)
--fs_license FS_LICENSE
Freesurfer license.txt
--coverage Run connectomemapper3 with coverage
--notrack Do not send event to Google analytics to report BIDS
App execution, which is enabled by default.
-v, --version show program's version number and exit
- Sebastien Tourbier (sebastientourbier)
- Yasser Aleman-Gomez (yasseraleman)
- Alessandra Griffa (agriffa)
- Adrien Birbaumer (abirba)
- Patric Hagmann (pahagman)
- Meritxell Bach Cuadra (meribach)
Collaboration Signal Processing Laboratory (LTS5) EPFL Lausanne
- Jean-Philippe Thiran
- Xavier Gigandet
- Leila Cammoun
- Alia Lemkaddem (allem)
- Alessandro Daducci (daducci)
- David Romascano (davidrs06)
- Stephan Gerhard (unidesigner)
- Christophe Chênes (Cwis)
- Oscar Esteban (oesteban)
Collaboration Children's Hospital Boston
- Ellen Grant
- Daniel Ginsburg (danginsburg)
- Rudolph Pienaar (rudolphpienaar)
- Nicolas Rannou (NicolasRannou)
Work supported by the Sinergia SNFNS-170873 Grant.
This software is distributed under the open-source license Modified BSD. See license for more details.
All trademarks referenced herein are property of their respective holders.
Copyright (C) 2009-2021, Hospital Center and University of Lausanne (UNIL-CHUV), Ecole Polytechnique Fédérale de Lausanne (EPFL), Switzerland & Contributors.