GCEx pipeline (August 2023)
Developed by Teymoor Saifollahi, Kapteyn Astronomical Institute
A data-analysis pipeline for the detection and characterization of extragalactic globular clusters (or generally star clusters).
(*This pipeline also makes use of several scripts/functions that have been developed by others)
Contact
Contact me in case of problems, questions, feedback, or to contribute to this pipeline:
- saifollahi@astro.rug.nl (this might not be available in the future, postdoc life)
- teymur.saif@gmail.com (personal email address)
Installation
In your working directory, execute the following command in the command line:
git clone https://github.com/teymursaif/GCTOOLS.git. You probably need to also install several Python libraries such as Astropy and photutils (latest versions) as well as packages Galfit (the executable is available in the repository), SExtractor and SWarp. you can install such libraries using pip install <library-name> in the command line (e.g. pip install astropy photutils).
Setup the input data
All the configuration of the pipeline is done in modules/initialize.py. Make sure that you have adjusted all the necessary inputs in modules/initialize.py. There are two kinds of inputs: the first group that you need to adjust based on your environment and study, and the second group that by default is set to a reasonable value and controls the work of the pipeline. You may leave them as they are. The most important parameters are:
WORKING_DIR: obviously, your working directoryFRAME_SIZE_ARCSEC: the dimension (in arcsec) of the area that you would like to analyzeTARGETS: your target(s) inside a list, where each component of the list is a string associated to a target and in this format:<some-ID> <some-given-name> <RA-in-deg> <Dec-in-deg> <distance-in-Mpc> <list-of-filters-separated-by-comma> <some-comments-about-the-target>.SE_executableandswarp_executable: the command that runs these applications (SExtractorandSWarp) in the command line.
Make sure data is prepared for the analysis by keeping them in the data_dir parameter as defined in modules/initialize.py. Frames for a given galaxy should be in the following format:
<galaxy-name>_<filter-name>.science.fits (for science frame), and <galaxy-name>_<filter-name>.weight.fits (for the associated weight-map, if available). Make sure you have PSF models stored in the psf_dir parameter as defined in modules/pipeline_functions. PSF model for a given filter should be given in the following format: psf_F606W.fits
Run the pipeline_functions
In your working directory and the command line execute: python gc_pipeline.py
Example
There is already data on MATLAS-2019, a UDG known for its rich GC population. After setting up the basics, you should be able to run the pipeline. Some output jpg files are already available in the output directory which you can look to get an idea of what to expect. There is data in 2 filters: HST/ACS in F606W and F814. There is more data on this object (but not included in the example)
Notes
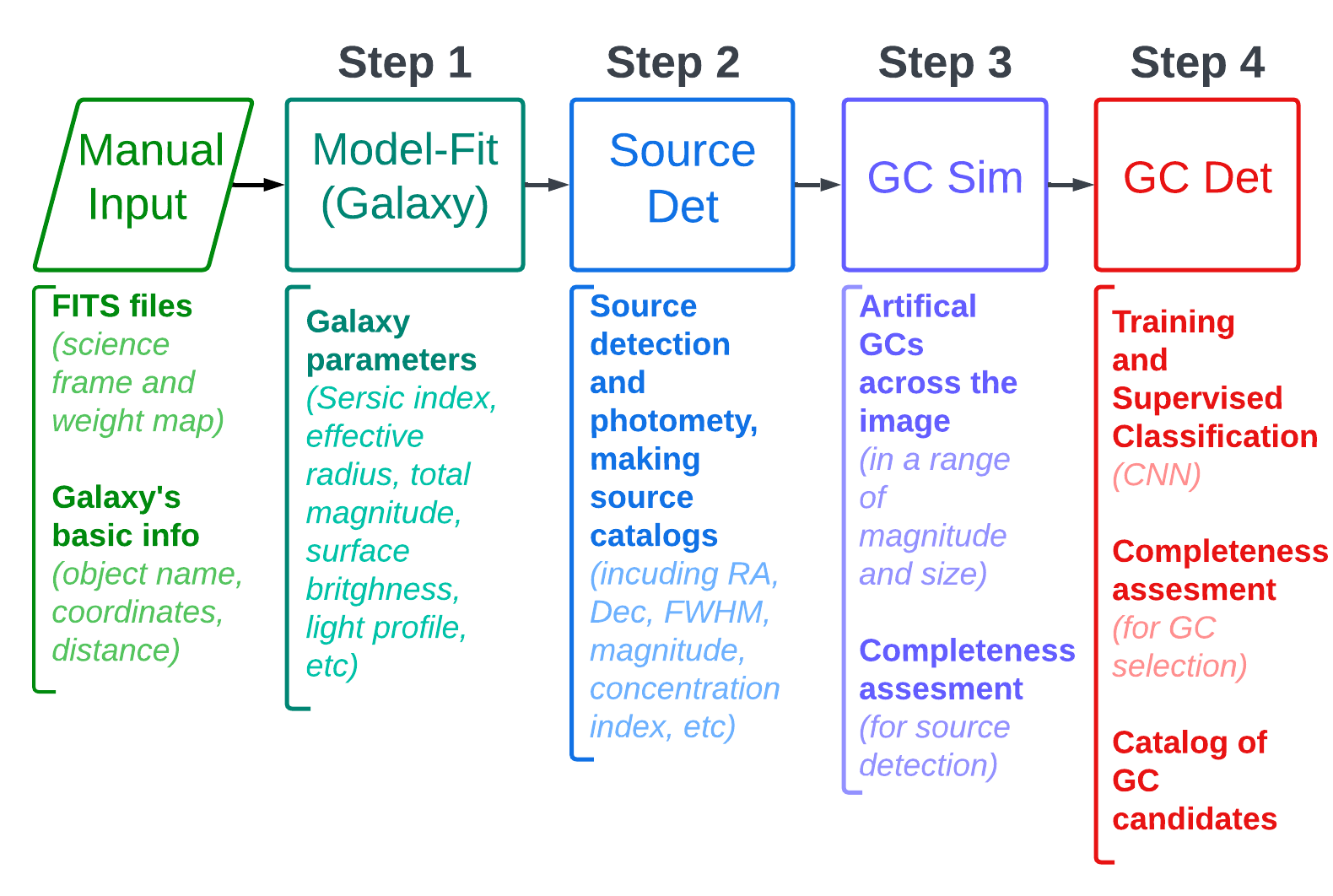
The current version (August 2023) of the pipeline does the first three steps of the desired analysis (out of 4 steps which will be available in the next versions). These two steps are (GCpipeline.png):
- Sersic modelling of the galaxy to estimate Sersic parameters of objects
- Source detection and photometry to make a source catalog with photometry in all filters
- Producing artificial GCs (at the distance of the galaxy and the given PSF) and measuring the completeness of source extractions
The 4th step (to be developed) is:
- Assesing the compactness index and colours of those imulated GCs, and GC selection criteria based on that and identifying GCs in the data, producing GC catalogs