Note : Please don't use it if you are not the fan of our biotrainee, Thanks.
source("http://bioconductor.org/biocLite.R")
install.packages('devtools')
BiocInstaller::biocLite("jmzeng1314/biotrainee")
library(biotrainee)But if you are in China, you should use the codes below:
options("repos" = c(CRAN="https://mirrors.tuna.tsinghua.edu.cn/CRAN/"))
install.packages("devtools",
repos="https://mirrors.tuna.tsinghua.edu.cn/CRAN/")
library(devtools)
source("https://bioconductor.org/biocLite.R")
options(BioC_mirror="https://mirrors.ustc.edu.cn/bioc/")
BiocInstaller::biocLite(c('airway','DESeq2','edgeR','limma'))
BiocInstaller::biocLite(c('ALL','CLL','pasilla','clusterProfiler'))
library(devtools)
source("https://bioconductor.org/biocLite.R")
options("repos" = c(CRAN="https://mirrors.tuna.tsinghua.edu.cn/CRAN/"))
options(BioC_mirror="https://mirrors.ustc.edu.cn/bioc/")
BiocInstaller::biocLite('org.Hs.eg.db')
install.packages("remotes",repos="https://mirror.lzu.edu.cn/CRAN/")
BiocInstaller::biocLite("jmzeng1314/biotrainee")
install.packages("pheatmap",repos="https://mirror.lzu.edu.cn/CRAN/")It will install many other packages for you automately, such as : ALL, CLL, pasilla, airway ,limma,DESeq2,clusterProfiler , that's why it will take a long time to finish if all of these packages are not installed before in your computer.
It always not very easy to download data if you are in China, so I also upload the file GSE42872_raw_exprSet.Rdata , you can load it directly.
if(F){
library(GEOquery)
gset <- getGEO('GSE42872', destdir=".",
AnnotGPL = F,
getGPL = F)
save(gset,'GSE42872.gset.Rdata')
}
load('GSE42872_eSet.Rdata')
b = eSet[[1]]
raw_exprSet=exprs(b)
group_list=c(rep('control',3),rep('case',3))
save(raw_exprSet,group_list,
file='GSE42872_raw_exprSet.Rdata')
Try to understand my codes, how did I filter the probes by the annotation of each microarry, and how I check the group information for the different samples in each experiment.
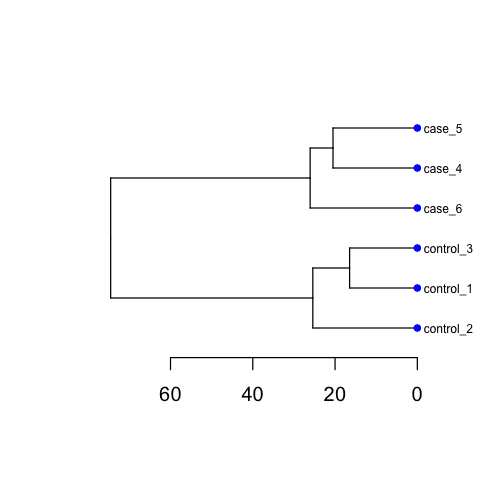
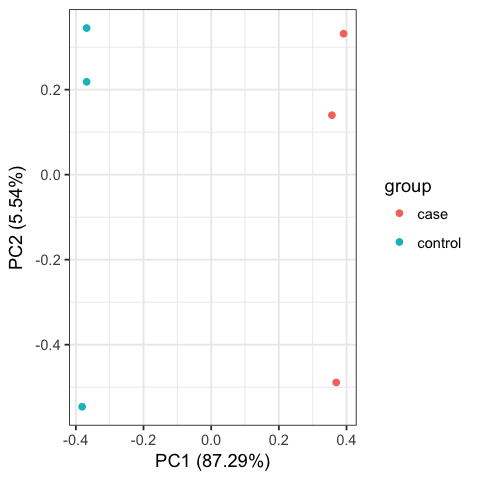
Including PCA and Cluster figures, as below:
Please ensure that you do run those codes by yourself !!!
Normally we will do differential expression analysis for the microarray, and LIMMA is one of the best method, so I just use it. If the expression matrix(raw counts ) comes from mRNA-seq, you can also choose DESeq based on negative binomial (NB) distributions or baySeq and EBSeq.
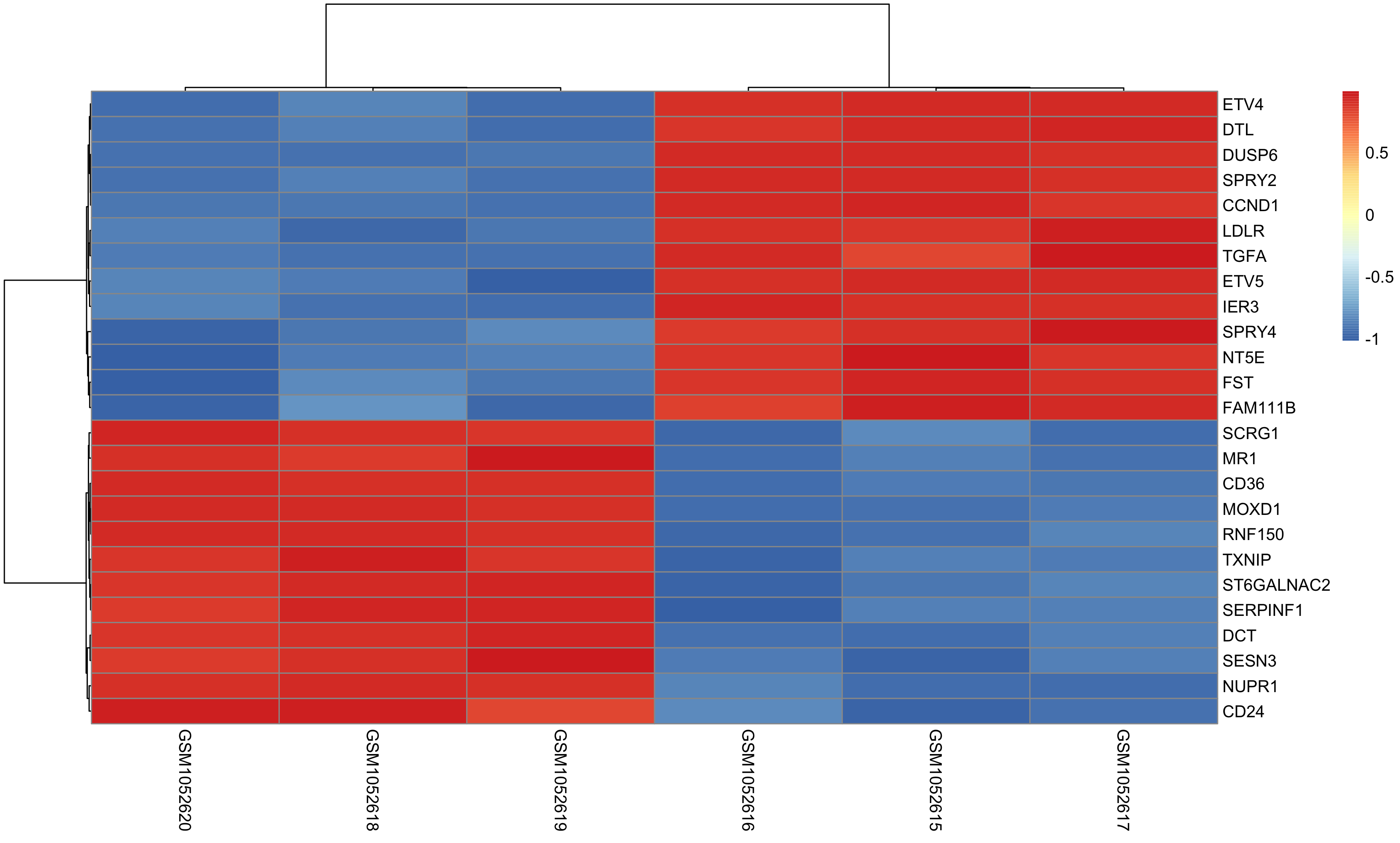
Once DEG finished, we can choose top N genes for heatmap as below:
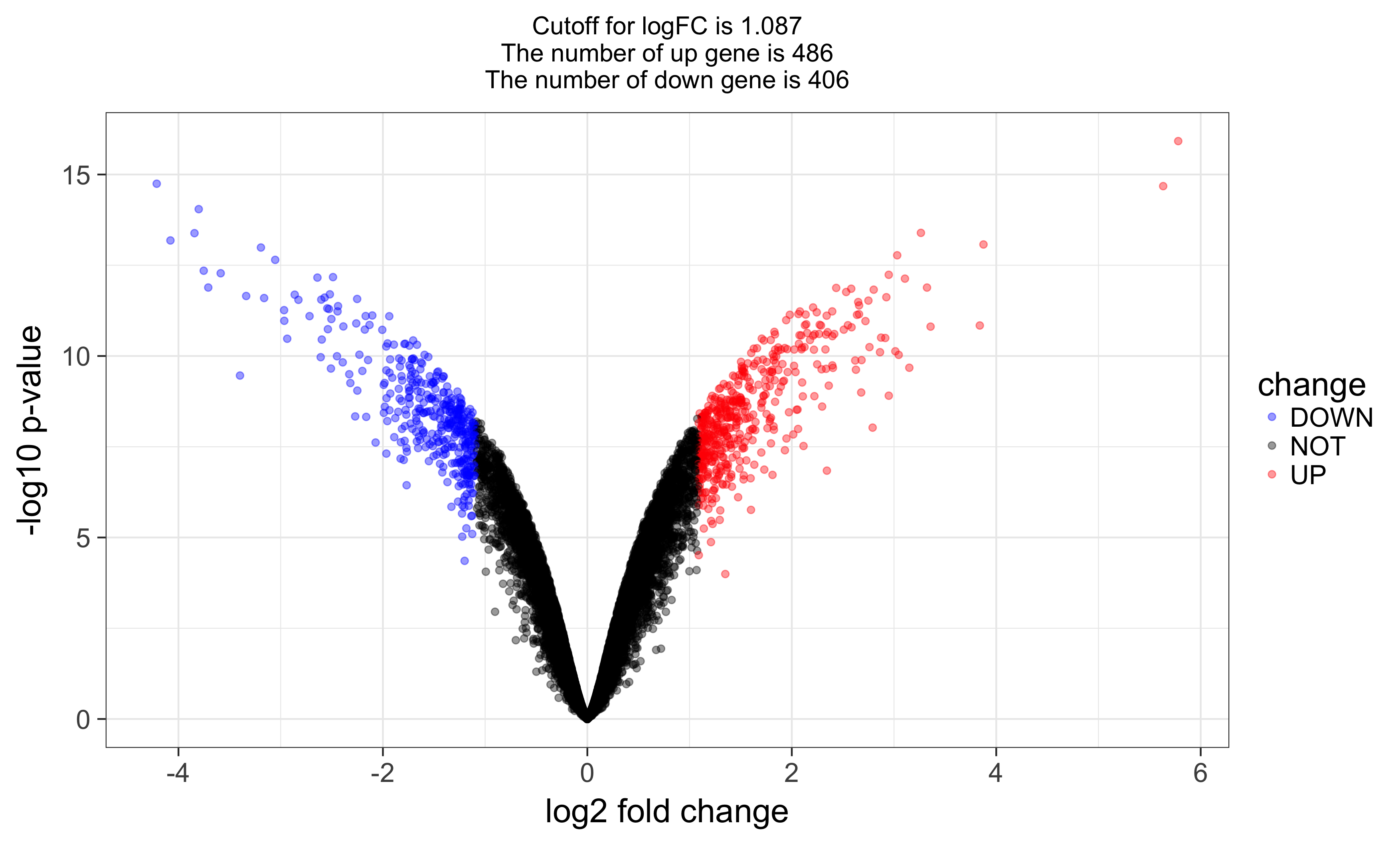
and volcano plot as below:
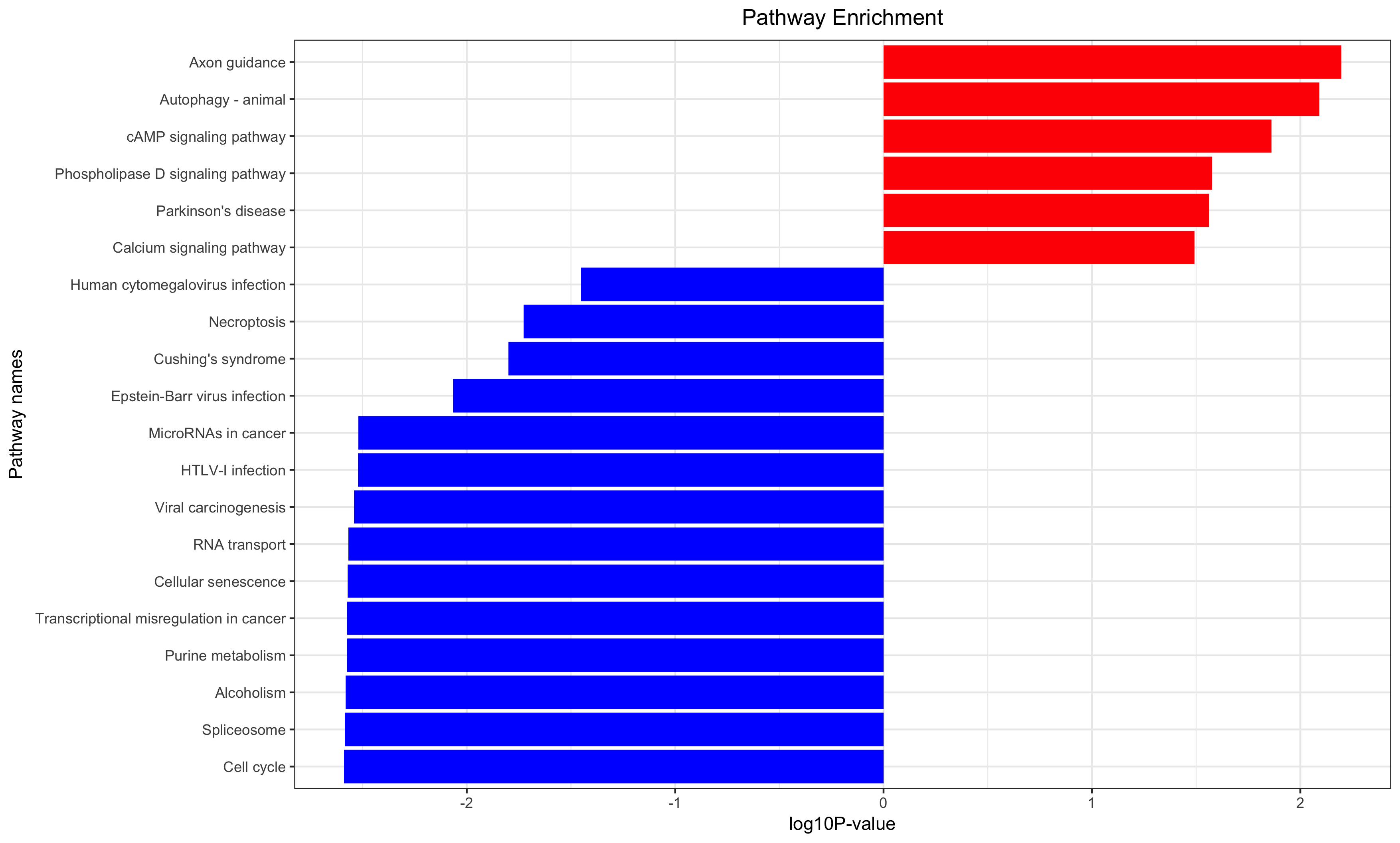
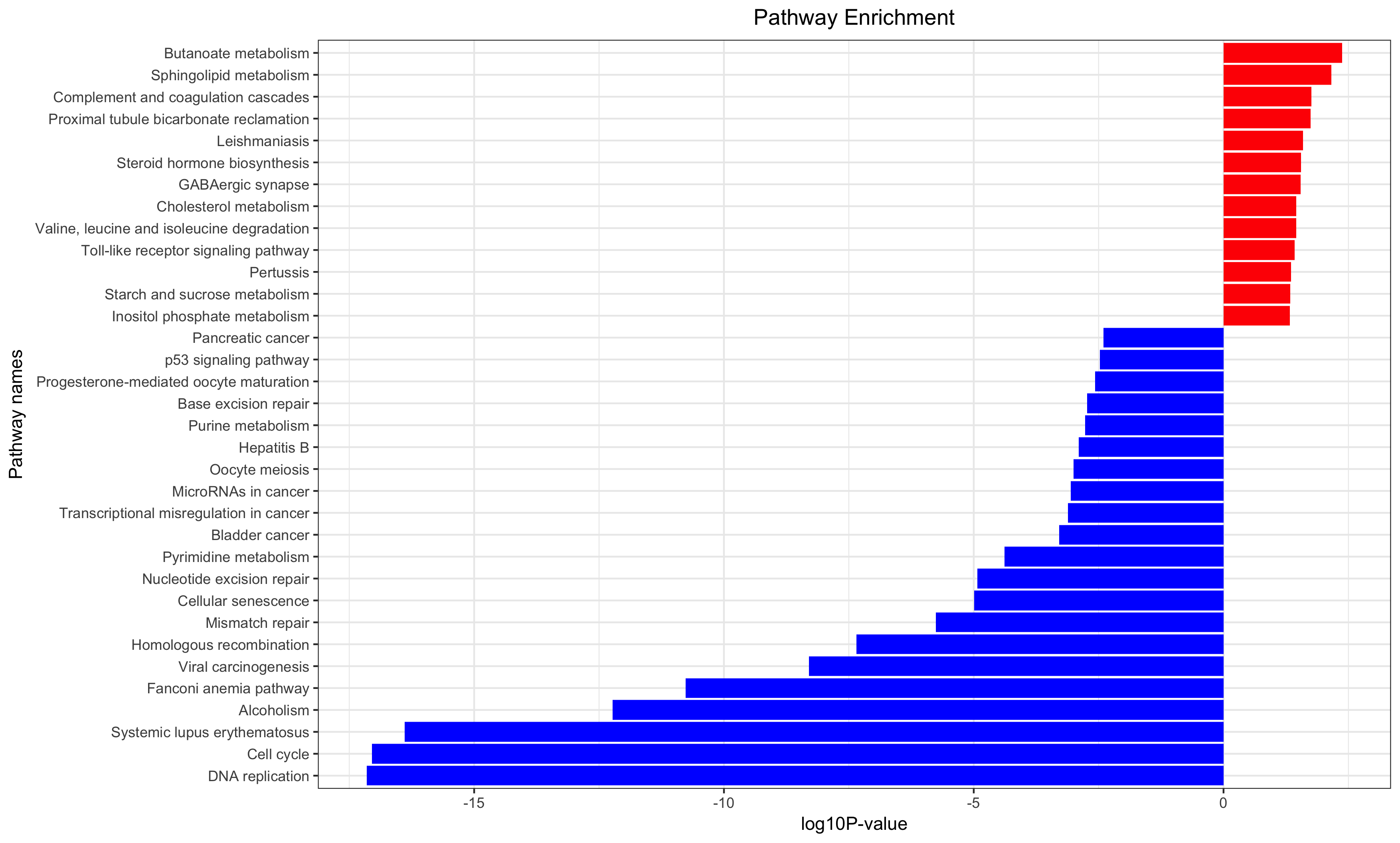
Annotation for the significantly changed genes, over-representation test or GSEA for GO/KEGG/biocarta/rectome/MsigDB and so on.
KM and cox
All the videos are uploaded in YouTube: https://www.youtube.com/channel/UC67sImqK7V8tSWHMG8azIVA/videos
如果你在中国,你可能会喜欢B站: https://www.bilibili.com/read/cv719181 ,视频链接: https://www.bilibili.com/video/av26731585/
其实不止是针对转录组表达芯片的数据分析教材,还有转录组数据处理流程,希望你可以仔细看,还有批量生存分析等各种其它统计分析方法我也会慢慢添加。
主要是根据大家的需求啦,希望大家多多反馈和提问哈!
如果你觉得我的教程对你有帮助,请赞赏一杯咖啡哦!
如果你的赞赏超过了50元,请在扫描赞赏的同时留下你的邮箱地址,我会发送给你一个惊喜哦!
关于我们
- 我的博客:生信菜鸟团 http://www.bio-info-trainee.com/
- 我们的论坛:生信技能树 http://www.biotrainee.com/thread-1376-1-1.html
- 我们的VIP社区:https://vip.biotrainee.com/d/311-
- 我们的微信公众号: https://mp.weixin.qq.com/s/egAnRfr3etccU_RsN-zIlg
- 我们的知识星球: https://t.zsxq.com/VjmQZNn
- 我们的腾讯课堂: https://biotree.ke.qq.com/
- 请善用搜索功能:http://weixin.sogou.com/
- 发邮件向我反馈 jmzeng1314@163.com