Prediction and characterization of T cell response by improved T cell receptors to antigen specificity with interpretable deep learning
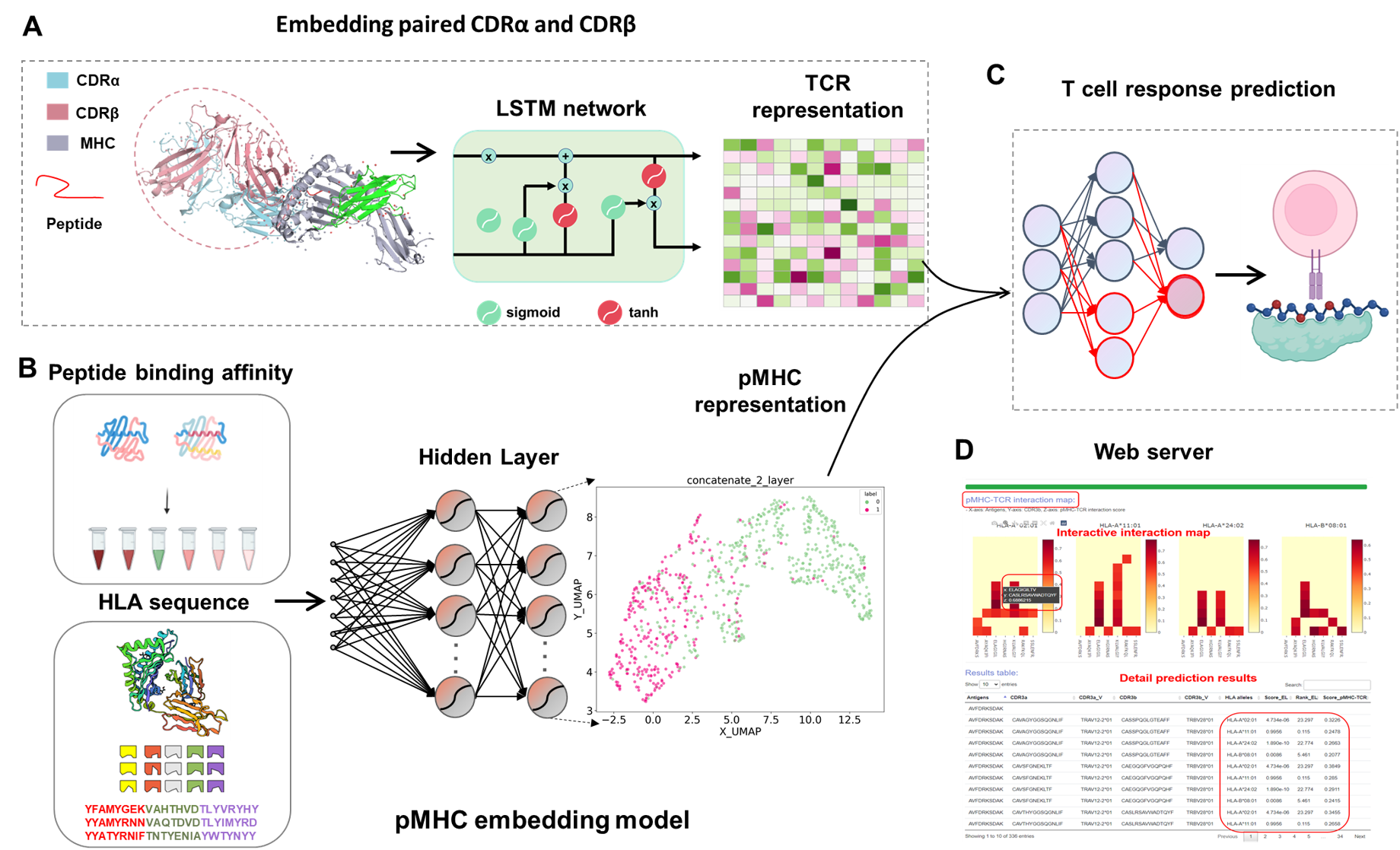
Cellular immunity is orchestrated by T cells through their immense T-cell receptors (TCRs) repertoire, which interact with antigenic peptides presented by major histocompatibility complex (pMHC) molecules, but the specificities of the T cell response is largely undetermined because of the huge variety of TCRs. Here, we present DeepTR, a one-stop collection of unsupervised and supervised deep learning approaches for pan peptide-MHC class I binding prediction, TCR featurization, and accurate T cell response prediction. DeepTR yields higher predictive performance and more efficient feature representation for peptide binding to MHC and enables superior antigen-specific TCR featurization than current state-of-the-art approaches. Through a transfer learning strategy, DeepTR provides accurate prediction of T cell activation achieved by mimicking crucial steps of the antigen presentation pathway. DeepTR also enables the discovery of specific TCR groups with a new regulatory mechanism and characterizes important contact residues that mediate TCR-antigen binding specificity. DeepTR may advance our understanding of the mechanisms of T cell-mediated immunity and yield new insight in both personalized immune treatment and development of targeted vaccines. DeepTR is freely available at https://bioinfo.uth.edu/DeepTR.
Download DeepTR by
git clone https://github.com/BioDataStudy/DeepTR.git
Installation has been tested in Linux server, CentOS Linux release 7.8.2003 (Core), with Python 3.7. Since the package is written in python 3x, python3x with the pip tool must be installed. DeepTR uses the following dependencies: numpy, scipy, pandas, h5py, torch, allennlp, keras version=2.3.1, tensorflow=1.15 shutil, and pathlib. We highly recommend that users leave a message under the DeepTR issue interface (https://github.com/BioDataStudy/DeepTR/issue) when encountering any installation and running problems. We will deal with it in time. You can install these packages by the following commands:
conda create -n DeepTR python=3.7
conda activate DeepTR
pip install pandas
pip install numpy
pip install scipy
pip install torch
pip install allennlp==0.9.0
pip install -v keras==2.3.1
pip install -v tensorflow==1.15
pip install seaborn
pip install shutil
pip install protobuf==3.20
pip install h5py==2.10.0
Please download large protein language model from the https://bioinfo.uth.edu/DeepTR/download/weights.hdf5. Note: right click - > save the link
Copy or move the weights.hdf5 file to the models\uniref50_v2\ directory to ensure that the model is preloaded successfully
Please cd to the folder which contains DeepTR_Prediction.py.
cd DeepTR_Prediction/
python DeepTR_Prediction.py -inf './data/test_tcr.txt' -out './prediction'
For details of other parameters, run:
python DeepTR_Prediction.py --help
test_tcr.csv: test csv file with 6 columns named as "CDR3a,CDR3a_V,CDR3b,CDR3b_V,Antigens,HLA_alleles": TCR-alpha CDR3 sequence, TCR-alpha V gene, TCR-beta CDR3 sequence, TCR-beta V gene, peptide sequence, and HLA allele.
DeepTR_predictions.tsv: result file includes the score of peptide binding to HLA and score of binding between pHLA and TCR.
Researchers can run DeepTR online at https://bioinfo.uth.edu/DeepTR. For commercial usage inquiries, please contact the authors.
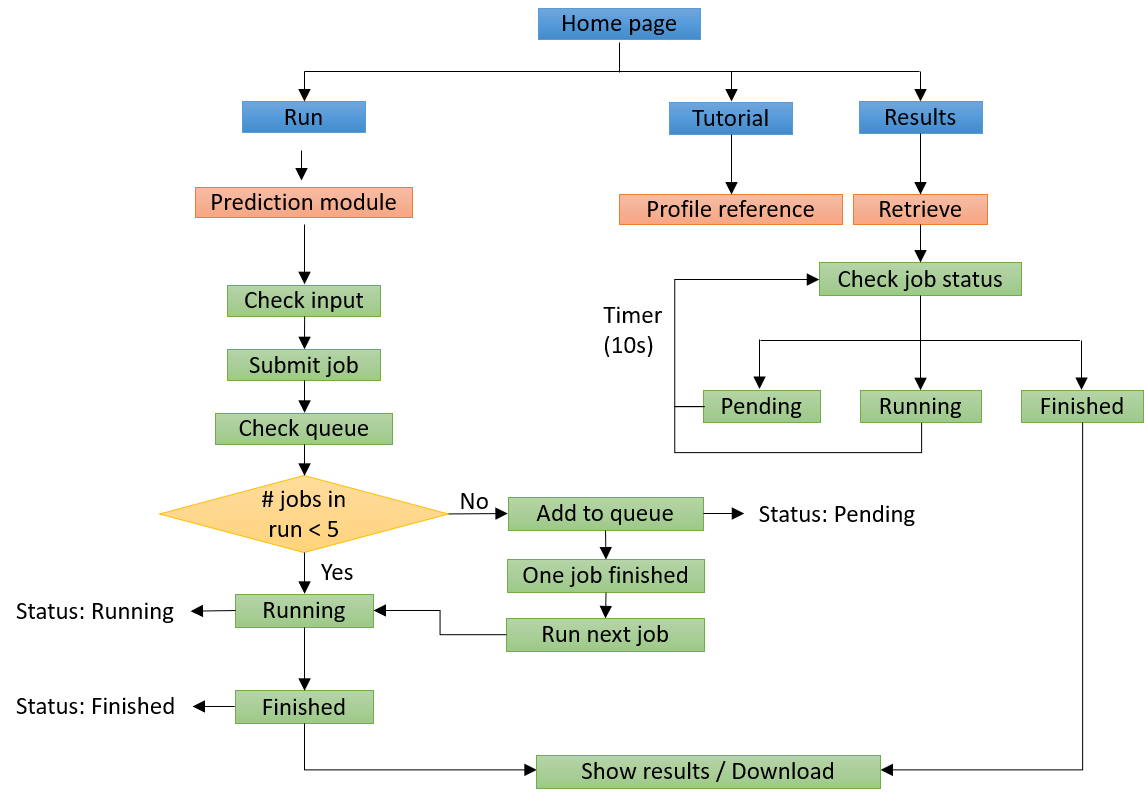
The workflow of the DeepTR online server is illustrated below. The information given is initially checked by the web server for proper formatting and TCR records, among other things. DeepTR launches a new job and changes the state of the work to "Running" after the input data is checked for correct format. Otherwise, DeepTR moves the new work to the bottom of the queue and changes its status to "Pending." Users are then routed to a website for tracking the job status after a job is successfully submitted, where the status is updated every 10 seconds until the task is complete. Through the unique job identifier, which can be generated automatically or customized by the submitter, users are able to monitor the job status and retrieve the result. .
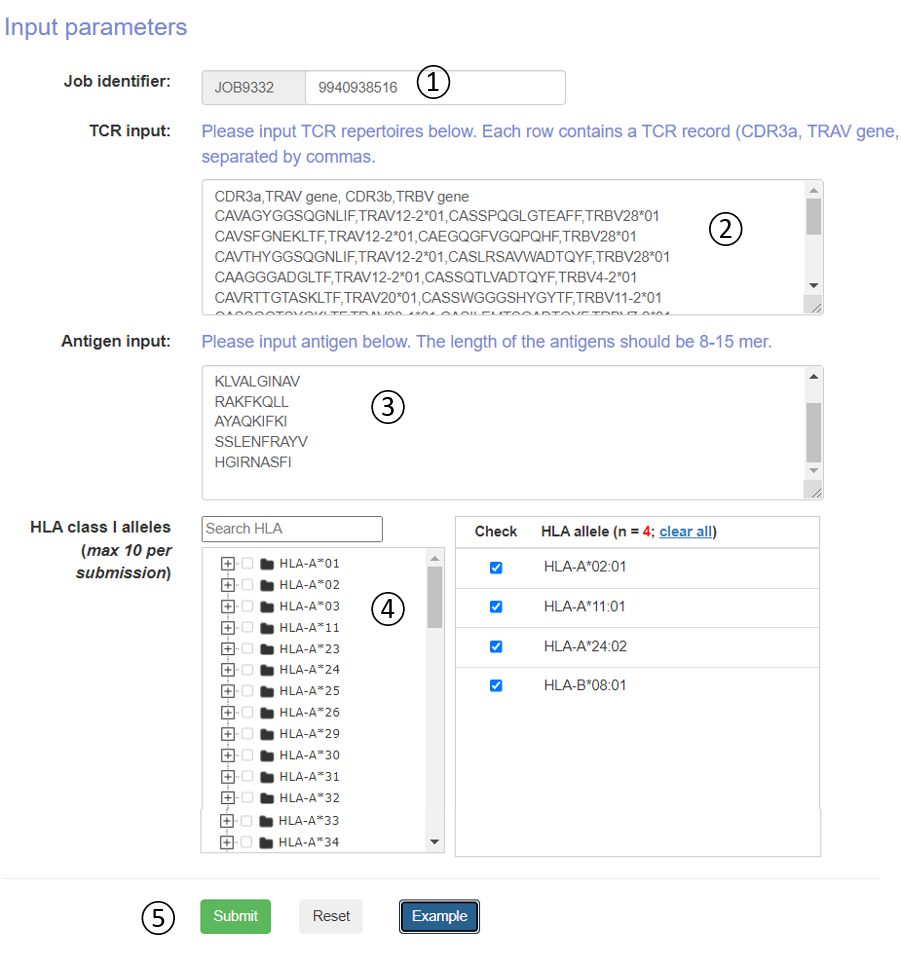
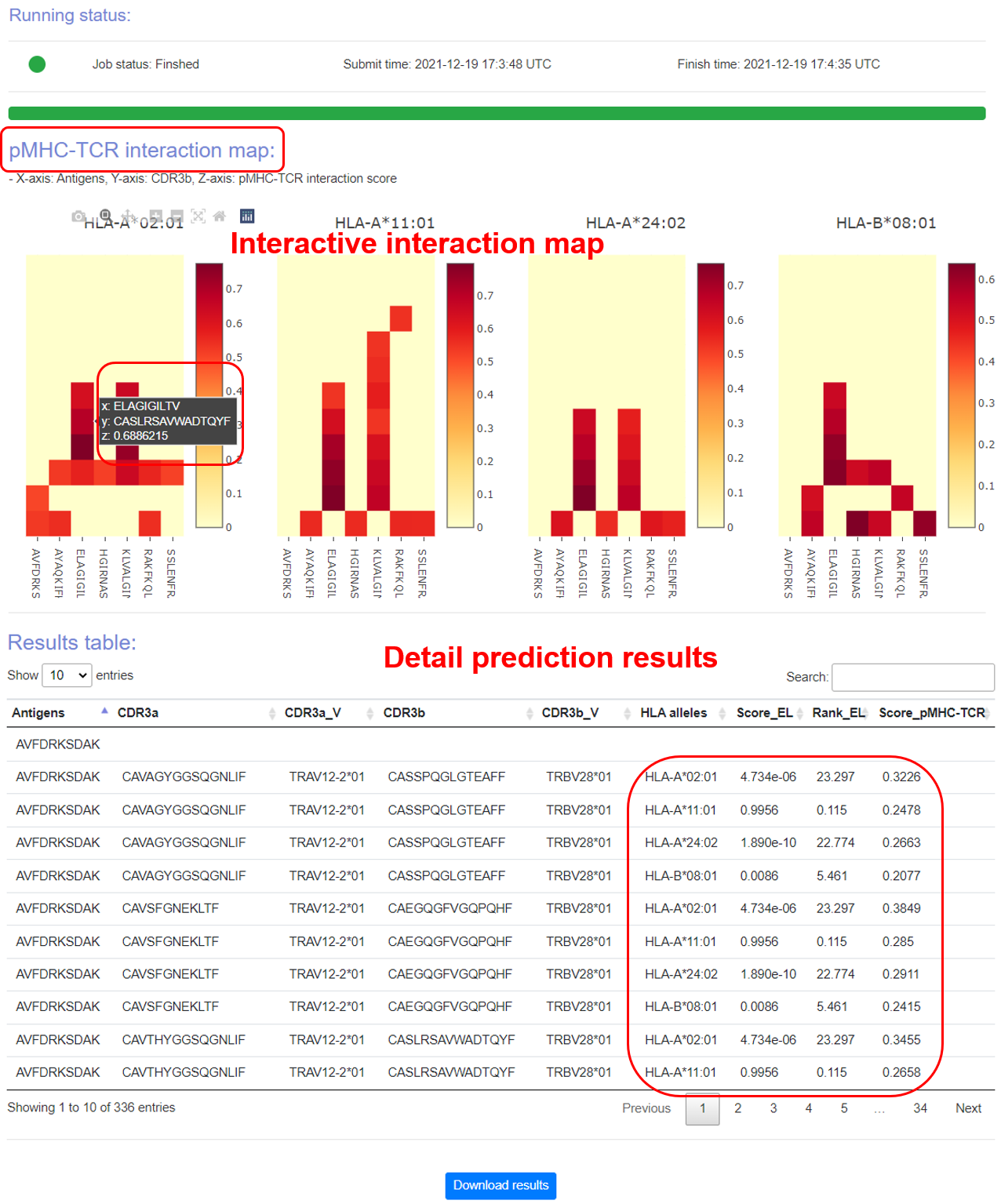
This online tool requires TCR repertoires through bulk or single-cell methods, including paired TCRα & β sequences of CDRs and V genes. Candidate antigens and HLA alleles also need be specified. The top ranked interaction between TCR and pMHC in each HLA allele would be visualized in an interactive way on the output page after a few minutes of running time. The whole prediction results, which are well organized in table format, could also be obtained, and downloaded. The testing has been conducted by multiple users from different countries. Tutorial is at https://bioinfo.uth.edu/DeepTR/Tutorial.php.
- Job identifier: Job identifier can be generated automatically or customized by the submitter. It is confidential to other users and can be used for job status monitoring and result retrieval.(See Results page).It is required.
- TCR input: Please input TCR repertoires below. Each row contains a TCR record (CDR3a, TRAV gene, CDR3b, TRBV gene) and separated by commas.
- Antigen input: The user can directly copy the antigens in the input box. The length of the antigens should be 8-15 mer.
- HLA alleles: The DeepTCR 1.0 server predicts pMHC-TCR binding to more than >100 well studied human MHC molecule. We constructed a classification tree of HLA. Users can quickly retrieve and submit candidate HLA alleles through the search box and tree map. Each submitted task is allowed to select up to 10 HLA alleles.
- Operation buttons: Submit, reset the submission form, or access the example dataset.
The top ranked interaction between TCR and pMHC in each HLA allele would be visualized in a interactive way on the output page after a few minutes of running time. The whole prediction results, which are well organized in table format, could also be obtained and downloaded.
Please cite the following paper for using: Xu H, Zhao Z. Prediction and characterization of T cell response by improved T cell receptors to antigen specificity with interpretable deep learning. In submission.