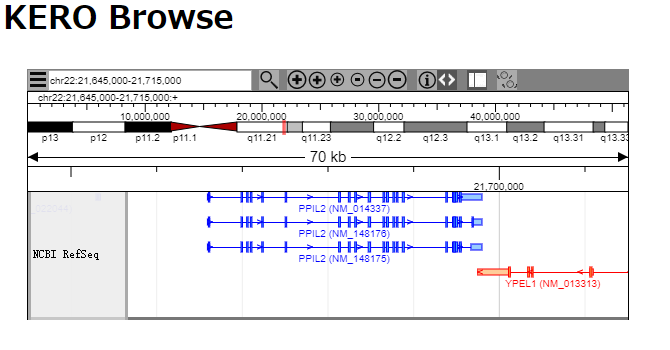
Simple HTML5 canvas based genome browser for NGS data. KERO-browse supports several file formats such as bam(+bai) or BigWig.
Web browser: Edge (V40 above), Google Chrome (V61 above) or Firefox (V56 above).
(JavaScript Generator supported web browser required)
For demo page installation:
cd /your/open/web_directory/
git clone https://github.com/DBKERO/genome_browser.git
#change proper file permission like:
#find ./genome_browser -type d -print | xargs chmod 755
#find ./genome_browser -type f -print | xargs chmod 644
#please access demo page: http://your_domain/genome_browser/sample.html
When you access demo page (http://your_domain/genome_browser/sample.html),
the following browser will be appeared:
Default genome version is UCSC hg38. The chromosome names are chr1, chr2..., chrM (not 1, 2..., Mt)
If you want to add track with your local PC files (supported only .bam+.bai or .bw), please select files as follows:
- Copy genome and RefSeq track data
cd /your/open/web_directory/genome_browser/test_data/
wget http://kero.hgc.jp/data/hg38/data.dat
wget http://kero.hgc.jp/data/hg38/genes.sorted.bb
#chmod if need
#Access to: http://your_domain/genome_browser/index.html
- Upload your bigWig file to server (e.g., /your/open/web_directory/genome_browser/test_data/foo.bw)
- Edit edit_me.js (genome_browser/js/edit_me.js)
- Add following line to edit_me.js at line 10:
var obigwig2 = new WgBigWig2("obigwig2", "#5555ff", "My bigWig", "test_data/foo.bw");
- Add ", obigwig2" to edit_me.js line 57.
//Show genome browser
gvObj.setupGenomeViewer(posStr, 1000, 300, [
"<hr /><div>Category buttons</div>",
"<div><input type=\"button\" id=\"show_item\" value=\"Show public data\" name=\"Sequence|Reference gene\" /></div>",
"<hr /><div>Public data</div>",
oseq, orefseq,
"<hr /><div>Your data</div>",
obigwig
]);
to
//Show genome browser
gvObj.setupGenomeViewer(posStr, 1000, 300, [
"<hr /><div>Category buttons</div>",
"<div><input type=\"button\" id=\"show_item\" value=\"Show public data\" name=\"Sequence|Reference gene\" /></div>",
"<hr /><div>Public data</div>",
oseq, orefseq,
"<hr /><div>Your data</div>",
obigwig, obigwig2
]);
- Access your web page (e.g. http://your_domain/genome_browser/index.html)
- Download fasta file of genomic sequences for target species to any directory (e.g. /your/open/web_directory/genome_browser/test_data/)
- Make indexed genome file to your web open directory (using genome_browser/scripts/get_indexed_genome.pl)
#In case of Chimpanzee in Ensembl
wget ftp://ftp.ensembl.org/pub/release-91/fasta/pan_troglodytes/dna/Pan_troglodytes.Pan_tro_3.0.dna_sm.toplevel.fa.gz
gunzip Pan_troglodytes.Pan_tro_3.0.dna_sm.toplevel.fa.gz
cd genome_browser/scripts/
#usage: perl get_indexed_genome.pl InputFasta OutPutPrefix
perl get_indexed_genome.pl ../../Pan_troglodytes.Pan_tro_3.0.dna_sm.toplevel.fa ../test_data/data
#chmod ../test_data/data.dat if need
-- get_indexed_genome.pl has been supported only less than 65536 sequences.
#In case of Chimpanzee in Ensembl(Pan_tro_3.0)
wget ftp://ftp.ensembl.org/pub/release-91/gtf/pan_troglodytes/Pan_troglodytes.Pan_tro_3.0.91.chr.gtf.gz
gunzip Pan_troglodytes.Pan_tro_3.0.91.chr.gtf.gz
cd genome_browser/scripts/
perl gtf2bed.pl ../../Pan_troglodytes.Pan_tro_3.0.91.chr.gtf ../test_data/data.chrom.sizes > ../test_data/genes.bed
sort -k1,1 -k2,2n ../test_data/genes.bed > ../test_data/genes.sorted.bed
#Please install Kent's bedToBigBed (For example: http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/)
wget http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/bedToBigBed; chmod 755 bedToBigBed
./bedToBigBed ../test_data/genes.sorted.bed ../test_data/data.chrom.sizes ../test_data/genes.sorted.bb
chmod 644 ../test_data/genes.sorted.bb
- Edit genome_browser/js/edit_me.js as follows:
Line 9-15, 57: Comment out the extra settings:
var obigwig = new WgBigWig2("obigwig", "#5555ff", "only chr22 ENCODE demo data", "test_data/ENCFF437TPA_chr22_cut.sorted.bw")
var urlSet = {
key2position: "https://dbtss.hgc.jp/cgi-bin/dbtss_autocomplete.cgi",
searchKey: "https://dbtss.hgc.jp/cgi-bin/keyword_search.cgi",
additionalParam: "SEE=1&UID=2&taxid=9606"
};
...
...
gvObj.setupGenomeViewer(posStr, 1000, 300, [
"<hr /><div>Category buttons</div>",
"<div><input type=\"button\" id=\"show_item\" value=\"Show public data\" name=\"Sequence|Reference gene\" /></div>",
"<hr /><div>Public data</div>",
oseq, orefseq,
"<hr /><div>Your data</div>",
obigwig
]);
to
//var obigwig = new WgBigWig2("obigwig", "#5555ff", "only chr22 ENCODE demo data", "test_data/ENCFF437TPA_chr22_cut.sorted.bw")
var urlSet = {
// key2position: "https://dbtss.hgc.jp/cgi-bin/dbtss_autocomplete.cgi",
// searchKey: "https://dbtss.hgc.jp/cgi-bin/keyword_search.cgi",
// additionalParam: "SEE=1&UID=2&taxid=9606"
};
...
...
gvObj.setupGenomeViewer(posStr, 1000, 300, [
"<hr /><div>Category buttons</div>",
"<div><input type=\"button\" id=\"show_item\" value=\"Show public data\" name=\"Sequence|Reference gene\" /></div>",
"<hr /><div>Public data</div>",
oseq, orefseq,
"<hr /><div>Your data</div>",
// obigwig
]);
Line 48: Set default display position:
var posStr = (loc[1])? loc[1]: "chr22:20900000-21000000";
to
var posStr = (loc[1])? loc[1]: "1:10000000-11000000";
Line 19-43: Delete and set chromosome sizes of your genome (see genome_browser/test_data/data.chrom.sizes)
var gvObj = new GenomeViewerObject("#viewer", {
"chr1": [1, 248956422],
"chr2": [2, 242193529],
...
...
"chrM": [25, 16569],
}, urlSet);
to
var gvObj = new GenomeViewerObject("#viewer", {
"1": [1, 228573443],
"2A": [2, 111504155],
...
...
"AACZ04055109.1": [44449, 984]
}, urlSet);
-- 1,2, ... 44449 are serial numbers.
- Edit index.html to proper content:
For example:
<title>Homo sapiens (UCSC hg38)</title>to<title>Pan troglodytes (Ensembl Pan_tro_3.0)</title>
<h1><i>Homo sapiens</i></h1>to<h1>Chimpanzee (<i>Pan troglodytes</i>)</h1>
Please access http://your_domain/genome_browser/index.html
See Preparation of your NGS result data
This project is licensed under the MIT License