This is for the course BIOL:4386: Intro to scientific computing in Spring 2021. This repository aims at following up C16 Nucleosome Occupancy Project and based on Bin He's repository C016-Pho4-binding_difference.
This project is unpublished and is originally from Dr. Bin He. Here is background, the biological question, the nature of the data, and the type of analyses I plan to perform.
Sacchromyces cerevisiae or S. cerevisiae, the baker's yeast, is closely related to the second-most common yeast pathogen Candida glabrata or C. glabrata. These two yeast species share over 90% 1-to-1 orthologs but occupy different ecological niches. This phenotypic difference is contributed tremendously by their distint gene regulatory networks. To get a better understanding of gene regulatory networks and how they evolve, we study phosphate starvation stress response (PHO), which is a well-established and typical model for studying stress responses. The main transcription factor involved in PHO is named Pho4. S. cerevisiae Pho4 or ScPho4 binds 74 promoter sites under phosphate starvation conditions; in contrast, C. glabrata Pho4 or CgPho4 binds to 115 promoter sites in the same genomic background (He et al, 2017). This difference is not due to factors, including the presence of concensus motifs CACGTG. Our hypothesis is CgPho4 is more capable of binding to nucleosome-occluded promoter sites compared with ScPho4.
This hypothesis is mainly motivated by a figure below. WHen deleting the non-consensus motif in the nucleosome-free region but keeping the consensus motif in the nucleosome-occluded site, it abolishes the expression of a target gene of Pho4, named PHO5. Pay attention to the two variants WT and A2. This observation provides a strong evidence of the importance of non-consensus motifs, and the huge impact of relative positioning of nucleosome occupancy and motifs on gene expression. CgPho4-specific sites (total of 115-74 = 41 sites) could show the similiar structual pattern like A2, in which only CgPho4 could access to the promoter while ScPho4 can not.
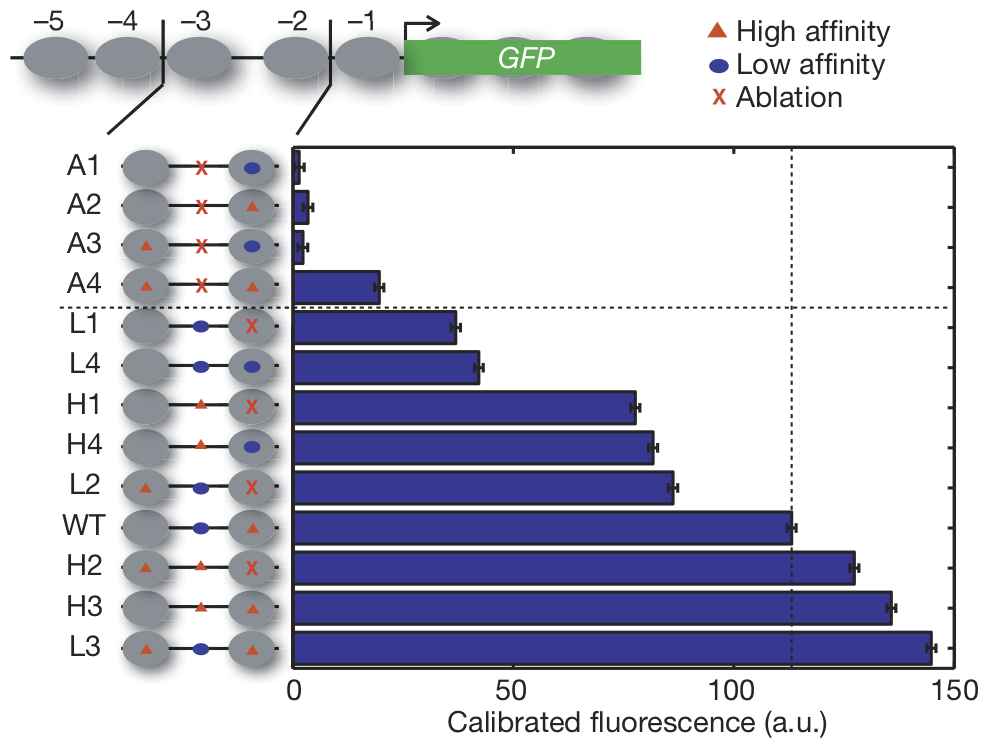 PHO5pr-GFP construct variants and expression under PHO. Grey circles are representative of nucleosomes. Red triangles stands for high affinity motifs CACGTG, while blue circles represent low affinity motifs, which are 1-bp off motifs (i.e. CACTTG). GFP is a reporter of gene expression.
PHO5pr-GFP construct variants and expression under PHO. Grey circles are representative of nucleosomes. Red triangles stands for high affinity motifs CACGTG, while blue circles represent low affinity motifs, which are 1-bp off motifs (i.e. CACTTG). GFP is a reporter of gene expression.
Why are some genomic sites only bound by *C. glabrata( Pho4 but not *S. cerevisiae( Pho4, in the same genetic background?
Determine if there are structural differences between the sites bound by both ScPho4 and CgPho4, vs those only accessible to CgPho4, in S. cerevisiae background.
| Data | Citation | Purpose |
|---|---|---|
| Nucleosome Occupancy | Zhou & O'Shea 2011 | To get the nucleosome occupany data; wig file through the GEO datasets or raw FASTQ files through the SRA Run selector |
| Binding sites of Pho4 | He et al 2017 | 115 sites for CgPho4; 74 sites for ScPho4 |
| Genome Sequence | SGD Website | the genome coordinates of the above table is based on S. cerevisiae genome R64-2-1, which is based on genome sequences of R64-1-1 |
(Need more advice)
- Mapping Promoter position as the X-axis
- Using RNA-seq data to produce nucleosome occupancy data from FAFSA file (see data). Use both high Pi (starting state before Pho4 takes effects) and low Pi conditions. Mapping the nucleosome occupancy data
- Finding and mapping Motif location.
- (Optional) Motif conservation within the sensu stricto species (the solid vs light shade in the PHO5 figure)
Lam, F., Steger, D. & O’Shea, E. Chromatin decouples promoter threshold from dynamic range. Nature 453, 246–250 (2008). https://doi.org/10.1038/nature06867
The expected figure would look like this. I plan to produce many figures to cover all binding sites of CgPho4 and ScPho4 under PHO to answer my biological question.
x axis is genomic coordinates of S. cerevisiae PHO5. Red triangles stands for high affinity motifs CACGTG, while blue circles represent low affinity motifs, which are 1-bp off motifs (i.e. CACTTG). The transparency is the conservity of motifs. Less transparent means more conserve. y axis measures the nucleosome occupancy. Higer y value means the higher nucleosome occupancy. The non-consensus motif (blue circle at -400) corresponses to a low nucleosome occupancy; in contrast, the red triangle (consensus motif) is under a nucleosome occupancy peak. This indicates this consensus motif is covered by a nucleosome.
