-
Notifications
You must be signed in to change notification settings - Fork 22
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Added fast scanning algorithm #5
Conversation
Codecov Report
@@ Coverage Diff @@
## master #5 +/- ##
=====================================
Coverage 100% 100%
=====================================
Files 3 5 +2
Lines 210 281 +71
=====================================
+ Hits 210 281 +71
Continue to review full report at Codecov.
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks good. The main thing I wonder about is code de-duplication---are there prospects for unifying the two implementations? Obviously there are a number of details (e.g., the use of two integers to left_label and top_label in one case and a Vector{CartesianIndex{N}} in the other), but I wonder if one could define some small utility functions that would allow you to combine the implementations?
This is not crucial, but the advantages of combining them include:
- more certainty that bugfixes will be applicable to all cases
- the potential for flexibility in allowing the usage of corners even in 2d
src/fast_scanning.jl
Outdated
| # for 2-D images | ||
| function fast_scanning{CT<:Colorant}(img::AbstractArray{CT,2}, threshold::Real, diff_fn::Function = default_diff_fn) | ||
|
|
||
| # Bfs coloring function |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
What does "bfs" stand for?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
By "bfs" I meant Breadth First Search
src/fast_scanning.jl
Outdated
| @@ -0,0 +1,235 @@ | |||
|
|
|||
| # for 2-D images | |||
| function fast_scanning{CT<:Colorant}(img::AbstractArray{CT,2}, threshold::Real, diff_fn::Function = default_diff_fn) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can we handle numbers too? CT<:Union{Real,Colorant} or something.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, that can be done!
src/fast_scanning.jl
Outdated
| end | ||
|
|
||
| # Remove labels that have been recolored due to the bfs calls | ||
| @noinline sort_fn!(removed_labels) = sort!(removed_labels) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I guess this is needed because of 15276?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, without it things lit up red in ProfileView.
src/fast_scanning.jl
Outdated
| _diagmN = diagm([1 for i in 1:N]) | ||
| half_region = ntuple(i-> CartesianIndex{N}(ntuple(j->_diagmN[j,i], Val{N})), Val{N}) | ||
| neg_neighbourhood{N}(x::CartesianIndex{N}) = ntuple(i-> x-half_region[i], Val{N}) | ||
| pos_neighbourhood{N}(x::CartesianIndex{N}) = ntuple(i-> x+half_region[i], Val{N}) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
So I guess one difference between the two implementations is that "corners" are used here but not above. I wonder if one could unify the two algorithms by having a AbstractStencilIterator, with different concrete implementations, and allowing that as an argument to the function?
|
After some more changes, the performance has increased with a mean time of about 40 ms for a 512x512 color image. default_diff_fn{CT1<:Colorant, CT2<:Colorant}(c1::CT1,c2::CT2) = sum(abs2,(c1)-Images.accum(CT2)(c2))
belongs_to(region, point) = default_diff_fn(region, point) < threshold^2 |
|
If unifying them is too painful, then by all means keep them separate. I am trying to head out for a holiday, but I'll see if I can review changes over the weekend. Sorry for the delay, I have been very busy with julia 1.0 changes lately... |
|
Seems that if a couple of comments above are addressed (spell out bfs and generalize to use |
|
Yes, currently I am working on replacing the |
9635237 to
31ab423
Compare
|
After removing the |
|
|
The author's implementation of the "Efficient Graph-Based Image Segmentation" paper post processes the image to remove small segments. They construct a Region Adjacency Graph with each of the segmented region as vertex and edges between neighboring regions measuring dissimilarity between these regions. They iterate through edges (sorted in increasing order of weight) and merge an edge's two vertices if either of the vertex regions has size < minimum region size. Since edge weights measure similarity between vertex regions, small regions will merge with their most similar neighbor (and this merged region will recursively merge with it's most similar region if it's size < minimum region size). Due to the way felzenszwalb's algorithm and normalized graph-cut algorithm are setup, their is no overhead for constructing the RAG and managing edge links during merge step. This step may slow down your algorithm. Choosing the similarity metric for the RAG that always works is also difficult. |
|
💯 for having succeeded in merging the two implementations with no loss in performance. You're leveraging what I think is Julia's main asset, the ability to write incredibly generic code without sacrificing speed. I think it's what sets Julia apart from anything I've ever used before, and you're becoming a master at exploiting this. I don't see anything that should hold this up from merging, unless you want to tackle the items in #5 (comment). Would the adaptive threshold produce a different value for each pixel? If so, could you once again stick with a single implementation, allowing an getscalar(A::AbstractArray, i) = A[i]
getscalar(a::Real, i) = aAs far as merging small components goes, one possibility would be to have a function that constructs a graph from a SegmentedImage keeping track of how many times each component is adjacent to each other component. This graph will be smaller than any pixelwise graph (since it only has as many nodes as there are labels, rather than pixels) and might allow quite a few different types of post-processing. |
|
I like the idea of allowing an |
|
Support for adaptive thresholding has been added. An array can be passed as |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks good! Merge at will.
| @@ -0,0 +1,136 @@ | |||
| sharpness{CT<:Images.NumberLike,N}(img::AbstractArray{CT,N}) = var(imfilter(img, Kernel.Laplacian(ntuple(i->true, Val{N})))) | |||
|
|
|||
| function adaptive_thres{CT<:Images.NumberLike,N}(img::AbstractArray{CT,N}, block::NTuple{N,Int}) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do you have a citation for this algorithm? Is it something that might have more widespread use?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do you have a citation for this algorithm?
The algorithm used in adaptive_thres has been demonstrated in this paper. They have used a linear combination of the variance and frequency as a measure of the threshold of each block. The coefficients have been adjusted to work with common grayscale images.
The sharpness (frequency) of an image is calculated using the variance of Laplacian as demonstrated here (see LAP4).
Is it something that might have more widespread use?
The sharpness function might be useful. However, there are multiple more methods for measuring the focus of an image (about 30 methods have been compared in the mentioned paper above). The adaptive_thres function uses constants that have been hard-coded to work nicely only with a small class of images and only when using abs(x1-x2) as the difference measure. I don't think it has much widespread use.
| """ | ||
| seg_img = fast_scanning(img, threshold, [diff_fn]) | ||
|
|
||
| Segments the N-D image using a fast scanning algorithm and returns a |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Come to think of it, a citation for most of our algorithms might be a good idea. I've not done that systematically in Images.jl, unfortunately.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I will add that
src/fast_scanning.jl
Outdated
|
|
||
| ``` | ||
| """ | ||
| fast_scanning{CT<:Images.NumberLike,N}(img::AbstractArray{CT,N}, block::NTuple{N,Int} = ntuple(i->4,Val{N})) = fast_scanning(img, adaptive_thres(img, block)) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Attach the docstring to the "main" method not the one that dispatches to the adaptive threshold?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also, wrap line after the =?
src/fast_scanning.jl
Outdated
|
|
||
| function fast_scanning{CT<:Union{Colorant,Real},N}(img::AbstractArray{CT,N}, threshold::Union{AbstractArray,Real}, diff_fn::Function = default_diff_fn) | ||
|
|
||
| if typeof(threshold) <: AbstractArray |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can also use isa(threshold, AbstractArray)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
or even threshold isa AbstractArray
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
threshold isa AbstractArray fails on Julia v0.5 but isa(threshold, AbstractArray) works. Do we need to support Julia v0.5?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Either is fine by me. If you want to drop 0.5 just make sure you change the REQUIRE and CI scripts appropriately.
|
🎆 |
| using Images, DataStructures, StaticArrays, ImageFiltering | ||
|
|
||
| # For efficient hashing of CartesianIndex | ||
| if !isdefined(Base.IteratorsMD, :cartindexhash_seed) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
is this needed if your minimum julia version is 0.6? nevermind, was added post-0.6 in JuliaLang/julia#22657 - Compat might be a better place for this
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Should I remove it from here and send a PR to Compat?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@timholy thoughts? Would this break anything in any way to start getting different hash values for these? If it's otherwise a performance-only improvement, would it make sense for this to be available in a centralized, approximately-always-loaded place instead of only being changed when ImageSegmentation is loaded?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It's just a performance tweak (though one with substantial consequences). I'd support moving it to Compat, though I don't think type piracy that leads only to performance changes is all that much to worry about. (Yes, better in Compat, but no crime having it only here.)

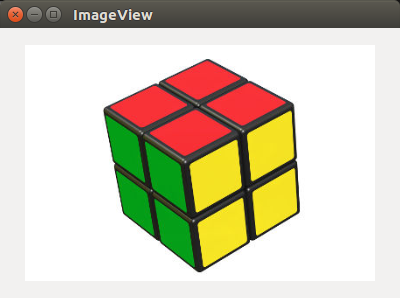
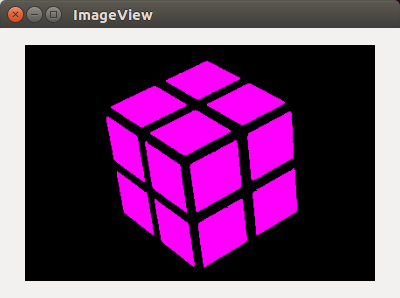
An implementation of the fast scanning algorithm as mentioned in this paper. An extension of this algorithm has also been implemented for working with higher dimensional images. A few features like removal of small segments and use of adaptive threshold will be progressively added. The performance of this algorithm for 2-d images is fantastic. After addition of the above-mentioned features, I will add the docstring and tests.
Here are a demo:
Original:
After segmentation and coloring the larger segments (except background):
Benchmark results: