This repository contains the code for our Scientific Reports 2024 Paper "Towards knowledge-infused automated disease diagnosis assistant".
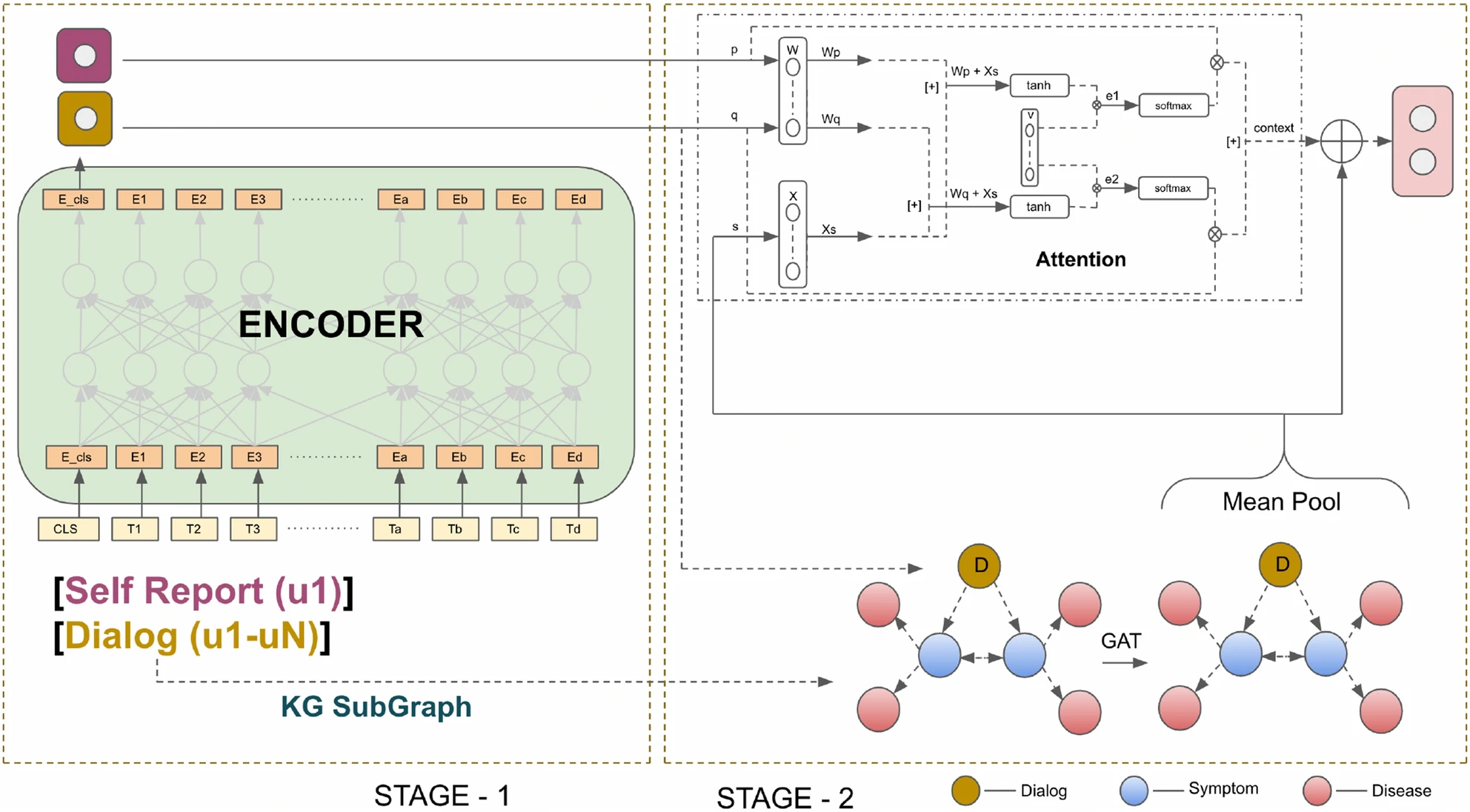
With the advancement of internet communication and telemedicine, people are increasingly turning to the web for various healthcare activities. With an ever-increasing number of diseases and symptoms, diagnosing patients becomes challenging. In this work, we build a diagnosis assistant to assist doctors, which identifies diseases based on patient–doctor interaction. During diagnosis, doctors utilize both symptomatology knowledge and diagnostic experience to identify diseases accurately and efficiently. Inspired by this, we investigate the role of medical knowledge in disease diagnosis through doctor–patient interaction. We propose a two-channel, knowledge-infused, discourse-aware disease diagnosis model (KI-DDI), where the first channel encodes patient–doctor communication using a transformer-based encoder, while the other creates an embedding of symptom-disease using a graph attention network (GAT). In the next stage, the conversation and knowledge graph embeddings are infused together and fed to a deep neural network for disease identification. Furthermore, we first develop an empathetic conversational medical corpus comprising conversations between patients and doctors, annotated with intent and symptoms information. The proposed model demonstrates a significant improvement over the existing state-of-the-art models, establishing the crucial roles of (a) a doctor’s effort for additional symptom extraction (in addition to patient self-report) and (b) infusing medical knowledge in identifying diseases effectively. Many times, patients also show their medical conditions, which acts as crucial evidence in diagnosis. Therefore, integrating visual sensory information would represent an effective avenue for enhancing the capabilities of diagnostic assistants.
Full Paper: https://www.nature.com/articles/s41598-024-53042-y
We provide the dataset for research and academic purposes only. To request access to the dataset, please follow the instructions below:
-
Fill Out the Request Form: To access the ES-MMD Dataset, you need to submit a request through our Google Form.
-
Review and Approval: After submitting the form, your request will be reviewed. If approved, you will receive an email with a link to download the dataset.
-
Terms of Use: By requesting access, you agree to:
- Use the dataset solely for non-commercial, educational, and research purposes.
- Not use the dataset for any commercial activities.
- Attribute the creators of this resource in any works (publications, presentations, or other public dissemination) utilizing the dataset.
- Not disseminate the dataset without prior permission from the appropriate authorities.
To train the model please follow the instructions
-
Make sure you are connected to GPU (Our experiments were carried out in Nvidia A100 GPU)
-
For training model run the file linear_plus_knowledge_self_report_having_first_utterance_one_disease.ipynb and save the weights corresponding to the best accuracy.
-
The code is for the case when each symptom has atmost one disease connected to them (one red node for each blue node in figure).
-
In one_disease_data you will find data that has 3 parts:
(a) final_combined_all_dialog.p - It has entire dialog.
(b) final_combined_all_self_report.p - It has only the very first utterance by patient i.e it has self report.
(c) joint_graph - It has the joint graph formed by combined subgraph and dialog node relevant to each dialog.
-
Results reported are mean value of running experiments 3 times.
If you find this code useful in your research, please consider citing:
@article{tomar2024towards,
title={Towards knowledge-infused automated disease diagnosis assistant},
author={Tomar, Mohit and Tiwari, Abhisek and Saha, Sriparna},
journal={Scientific Reports},
volume={14},
number={1},
pages={13442},
year={2024},
publisher={Nature Publishing Group UK London}
}For any queries, feel free to contact Mohit Tomar (mohitsinghtomar9797@gmail.com)