-
Notifications
You must be signed in to change notification settings - Fork 0
Model of Ocean Biogeochemistry and Isotopes (MOBI)
The Model of Ocean Biogeochemistry and Isotopes (v2.1) is a unique tool for climate and ocean research. It simulates cycles of carbon, nutrients and isotopes and includes relatively simple plankton food web dynamics. It has been coupled to different physical models such as the UVic model, the Transport Matrix Method (TMM) and Veros.
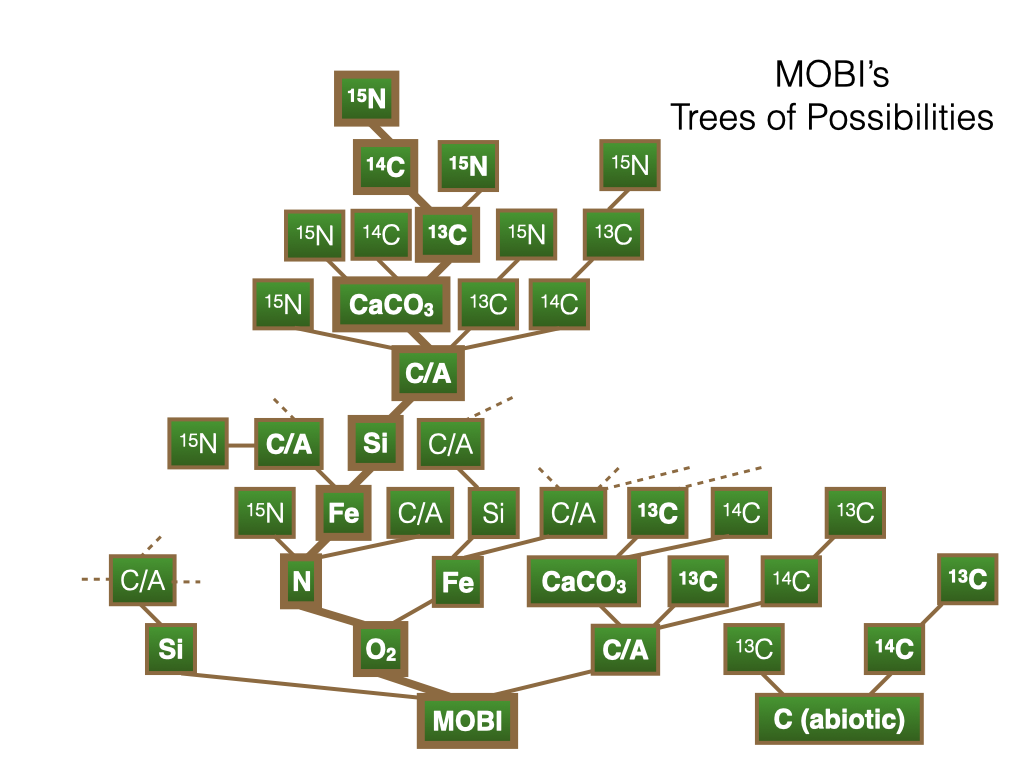
MOBI can be run in many different configurations as illustrated here. The most basic version is a simple NPZD model with one nutrient, one phytoplankton, one zooplankton and one detritus type. This version has 4 prognostic tracers and is fast, whereas the most complex and computationally expensive version has 33 prognostic tracers including three phytoplankton types, four inorganic nutrients (nitrate, phosphate, silicate and iron), dissolved organic matter, and isotopes of nitrogen and carbon. Many different intermediate configurations have also been tested as shown in bold font in MOBI's tree of possibilities.
Different configurations can be created by switching on/off certain options. Some options depend on other options. E.g. nitrogen isotopes (15N) require the nitrogen option to be switched on. Nitrogen requires oxygen to calculate denitrification. Carbon and alkalinity (C/A) should be switched on together for a fully interactive carbon cycle. However, the carbon cycle can be run without biology, which results in an inorganic carbon model (small separate tree on the right). Thus, each researcher or group can run a custom configuration.
MOBI works with C++ preprocessor commands that start with # if defined OPTION statements, which can be used to switch on and off parts of the code in the *.F files. Compiling the UVic model involves mk e, which uses options in the mk.in file to select code and copy it into the \code subdirectory. Currently there are the following MOBI related options available.
O_mobi
O_mobi_alk
O_mobi_caco3
O_mobi_o2
O_mobi_nitrogen
O_mobi_nitrogen_15
O_mobi_silicon
O_mobi_iron
#O_mobi_no_vflux
#O_save_mobi_fluxes
#O_save_mobi_diagnostics
#O_mtlm_carbon_13
#O_mtlm_carbon_14
O_carbon
#O_carbon_co2_2d
#O_carbon_uncoupled
#O_carbon_13
#O_carbon_13_coupled
#O_carbon_14
#O_carbon_14_coupled
In the above example from the mk.in file the options starting with a pound sign (#) are commented out and thus deactivated.
MOBI code is contained in two files: mobi_src.F and mobi.h.
mobi.h contains indices and parameters shared by different routines. An important parameter is the number of prognostic tracers ntnpzd.
parameter (ntnpzd = 4 ! po4, phyt, zoop, detr
#if defined O_carbon
& + 1 ! dic
The code snippet above shows that the basic model version (with only O_mobi switched on) includes 4 prognostic tracers. Switching on O_carbon adds DIC (Dissolved Inorganic Carbon) as one additional tracer.
MOBI is structured as a vertical column model with three subroutines in mobi_src.F.
-
mobi_initinitializes parameters and indices -
mobi_driverprepares and calls mobi_src for the whole water column -
mobi_srccalculates sources and sink terms for one vertical grid box
In UVic mobi_init is called by set_mom.
:
call setimobi('imobiX',imobiX,imobi)
:
The call to setmobi initializes the tracer name indices (imobiX) used to identify the tracers. E.g. call setimobi('imobidin15',imobidin15,imobi) sets the index for nitrogen-15 in dissolved inorganic nitrogen. Each call to setimobi increments imobi by one for use in the next call.
In UVic called by tracer.
:
do k=1,kmx ! this is the main loop over the vertical gridboxes
:
impo = expo*dztr(k) ! import of organic matter into box k is the export from box k-1
call mobi_src(tnpzd, impo, ..., snpzd, expo) ! calculation of source terms for each box
snpzd = snpzd*rdtts ! rdtts = 1/dtts divide by tracer timestep to get the tendencies
expo = expo*rnbio ! rnbio = 1/nbio divide by number of bio timesteps to get average
bdeni = ... ! calculate benthic denitrification
expo = expo - sgb_in(k)*expo ! loss to subgrid sediments
src(k,isX) = snpzd(imobiX) ! pass source terms to UVic
prca = ... ! calcite production
expo = expo*dzt(k) ! multiply with level thickness for next layer import
:
enddo
do k=1,kmx
:
wcdeni = ... ! water-column denitrification
:
enddo
do k=1,kmx
:
... ! calcite and opal calculations
:
enddo
Tracer concentrations are stored in the tnpzd(k,imobiX) array, whereas source and sink terms are stored in the snpzd(imobiX) array. Tracer concentrations in UVic (t(i,k,j,iX,tau)) use the iX index to identify tracers. Thus exchange between MOBI and UVic can use statements such as this tnpzd(:,imobipo4) = t(i,:,j,ipo4,taum1) for phosphate. Source terms in UVic (src(k,isX)) use isX and can be exchanged like src(k,ispo4) = snpzd(imobipo4).
Called by mobi_driver.
:
bioX = bioin(imobiX)
do n=1,nbio
:
bioX = bioX + dtbio*(SMSX) ! update tracer concentrations
:
expoout = expoout + expo
enddo
bioout = bioX - bioin(imobiX)
The routine receives input variables such as bioin and impo from mobi_driver. After some preparatory calculations it loops over nbio biological time steps. During each time step source-minus-sink fluxes (SMSX) are calculated for each biological variable and applied to update the concentrations. Export fluxes are accumulated to be passed on to the next level in mobi_driver.
Several UVic files and routines are associated with MOBI-UVic coupling.
:
tnpzd() = ...
call mobi_driver(..., tnpzd,..., src())
source() = src()
:
t() = t() + twodt*(transport + source())
:
The UVic model subroutine tracer calculates light, tnpzd and other input variables and passes those to mobi_driver, which returns the source-minus-sink terms src. Those are then used together with the transport terms to update all tracers.
Initialize ocean model.
:
call setmobi
:
Get ocean surface boundary conditions.
:
sbc(i,j,iXflx) = sbc(i,j,iXflx) + gaost(iX)*tmp
:
Calculation of surface fluxes including virtual fluxes.
Get atmospheric surface boundary conditions.
:
call data (is, ie, js, je) ! get atmospheric boundary conditions
call co2calc_SWS
sbc(i,j,idicflx) = ...
co2ccn = co2ccn + ...
sbc(i,j,io2flx) = piston_o2*(o2sat - sbc(i,j,isso2))
:
Calculation of air-sea carbon and oxygen fluxes and atmospheric CO2.
:
call get_tdsbc (...) ! interpolates time-dependent forcing data (e.g. iron fluxes) to current time
:
sbc(i,j,index) = wprev(n)*obc(i,j,n,iprevm(n))
& + wnext*obc(i,j,n,inextm(n))
Size parameters for tracer arrays.
:
nt = ...
nsrc = ...
:
Common blocks include indices for tracer arrays.
:
integer iphyt, izoop, idetr, io2, ino3, idiaz, idop, idon
common /mw_i/ ipo4, iphyt, izoop, idetr, io2, ino3, idiaz
:
integer istemp, issalt, isdic, isc14, isalk, ispo4, isphyt ! surface
common /mw_i/ istemp, issalt, isdic, isc14,isalk, ispo4, isphyt
:
Main program.
:
call set_mom
call setdata ! read boundary conditions from files in /data
do n=1,numseg ! main time loop over numseg segments
call gasbc (1, imt, 1, jmt) ! get atmospheric boundary conditions
:
call gosbc (1, imt, 1, jmt) ! get ocean boundary conditions
do loop=1,ntspos ! run ocean model ntspos times per segment
call mom
:
enddo
enddo
:
Main time stepping loop calls gasbc, gosbc and mom.
Main ocean model.
:
call setvbc (joff, js, je, is, ie)
call tracer (joff, jstrac, jetrac, is, ie)
:
Calls setvbc and tracer.
Set vertical boundary conditions.
:
stf(i,j,iX) = ... ! surface tracer flux
btf(i,j,iX) = ... ! bottom tracer flux
:
Most input and output (IO) in UVic is through netcdf files. Input files are in the /data subdirectory whereas output files (tsi and tavg) are written to the main directory. Timeseries indices such as global average temperature and circulation indices are written to the tsi file, which is small and thus can be written frequently (e.g. every 10 days). Time averages of the full 3-D fields (tavg file) are large and are typically written less frequently to save space.
Some tracers require external input files. E.g. iron requires maps of surface iron fluxes associated with dust input (O_feflux.nc) and bottom fluxes associated with hydrothermal activities (O_fe_hydr.nc). Since hydrothermal fluxes don't depend on time they can be read in just once at the start of the run in mobi_init. Aeolian iron fluxes, on the other hand, depend on the month of year and thus are read in data using get_tdsbc(..., s, o), which interpolates in time and allows for unit conversion by using a scalar (s) to multiply the input data and and offset to add to the input data (in this case there is no unit conversion since s=1 and o=0).
:
call get_sbc (...) ! initializes monthly-climatology ocean surface boundary conditions from data (e.g. iron fluxes)
:
obc() = ... ! obc is used in data (see above)
Netcdf output is done in two files: mom_tsi.F and mom_tavg.F. Both use netcdf routines such as defvar and putvara from uvic_netcdf.F. defvar is used to define variable names, units, axes and such, whereas putvara writes the variables to the netcdf file.
:
call defvar ('O_dic', iou, 4, it, -c1, c100, ' ', 'F'
&, 'dissolved inorganic carbon', ' ', 'mol m-3')
:
:
tmpijk(ils:ile,jls:jle,kls:kle) = t(ils:ile,jls:jle,kls:kle,n)
:
call putvaramsk('O_dic', iou, ln, ib, ic, tmpijk, tmpijkm
&, c1, c0)
: