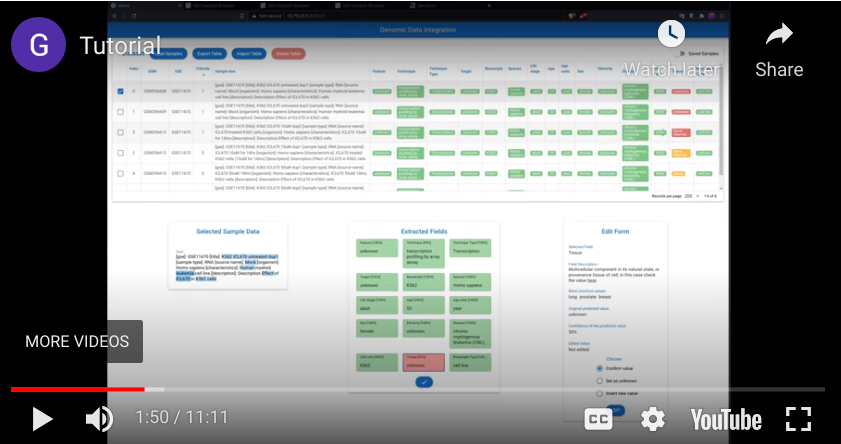
Welcome to the Genomic Metadata Integration tool! with this web-tool is possible to extract structured tables from the metadata of all the experiments contained in the Gene Expression Omnibus.
- Install the last version of Docker
- Download the containers with the commands:
docker pull 2603931630/gemi:frontenddocker pull 2603931630/gemi:backend - Download the repository:
git clone https://github.com/armando2603/gemi - Install the nodes.js dependencies:
docker run --rm -it -v "/$(pwd)/gemi/frontend/:/usr/src/app/" -p 51111:8080 2603931630/gemi:frontend npm install - Download the pretrained model at this link drive
- Copy the downloaded file in the background/Models/ folder
- Run the containers:
docker run -d --rm -it -v "/$(pwd)/gemi/frontend/:/usr/src/app/" -p 51111:8080 2603931630/gemi:frontend quasar dev && docker run -d -v "/$(pwd)/gemi/backend/:/workspace/" -p 51113:5003 -it --rm --gpus all 2603931630/gemi:backend flask run -p 5003 -h 0.0.0.0 - In the browser go to the page http://localhost:51111/#/
Remember that you need a gpu NVIDIA in your machine