Recan_gui is a Django-based web version of recan. It allows to analyze recombination events in viral genomes using genetic distance plotting. The tool is currently available at http://yuriyb.pythonanywhere.com.
Input files should be multiple sequence alignments in fasta format (with extenstions: fasta, fas or fa).
To upload an alignment click: choose file button and then upload your alignment button. After alignment is uploaded, the list of sequences it contains will appear on the left.
potential recombinant. It's chosen from the list of sequences. All the distances will be plotted relatively to this sequence.window size. Sliding window size in nucleotides. It determines the 'sequence slice' which we take to calculate genetic distance.window shift. Sliding window shift in nucleotides. It determines how fast we move the window along the array of sequences. It must be less than window size. The smaller is the shift the more detailed is the plot.region startandregion end. If you need to examine a particular genomic region more closely, you can adjust these values. Window will roll along the alignment fromregion starttoregion end. In this case only data from the given region will be plotted. When you're happy with your settings hitplotbutton.
When the plot is constructed, hover your mouse over the top-right corner of the plot. It shows the plot control menu. The menu allows to drag, resize the plot and download the screenshot. The sequences with their color traces are shown above the plot. You can click on any of them to hide or show a chosen trace. You can also double click on any sequence legend to isolate the single trace or return all the traces to the plot.
If you didn't use the distance method for virus recombination study before, you can download example datasets and play with them first. Each dataset contains sequences of a particular virus (HBV, HIB, HCV, LSDV, norovirus). Among the other sequences there is a recombinant virus and its major and minor parents in these alignments (except for the file hbv_C_Bj_Ba.fasta, it contains only recombinant and its parents). Minor parents have 'min' ending in their sequence name. Major parents have 'maj' ending. Recombinant sequences are designated with 'rec' ending. All the recombinant viruses included in the articles are described in the articles (see references below).
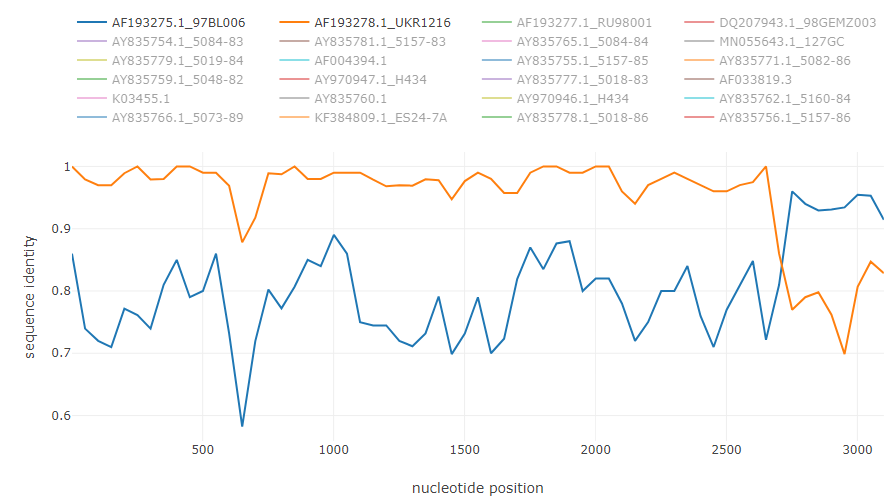
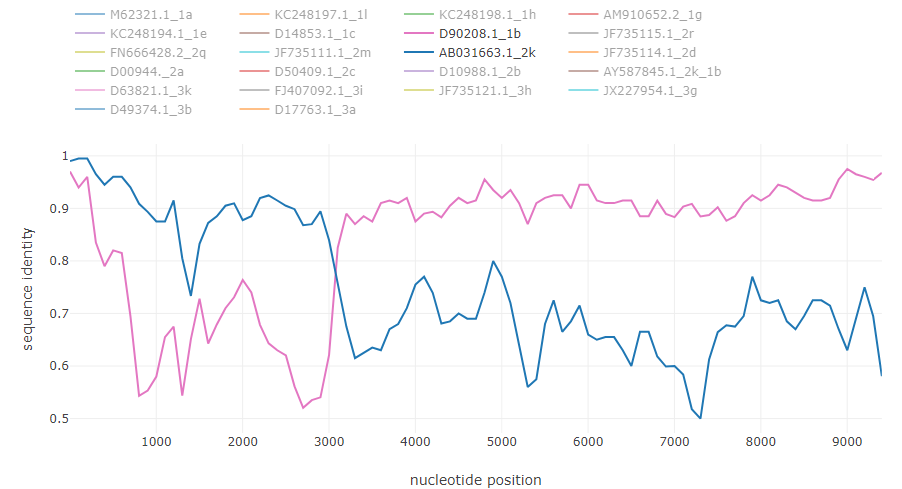
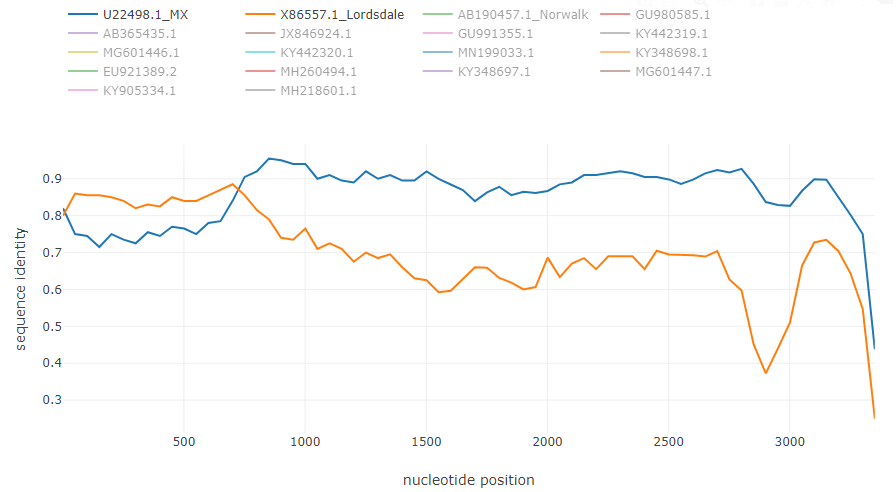
Typical recombination events from the example
HCV intergenotype recombinant 2k/1b:
Norovirus recombinant isolate:
- Hepatitis B Virus of Genotype B with or without Recombination with Genotype C over the Precore Region plus the Core Gene. Fuminaka Sugauchi et al. JOURNAL OF VIROLOGY, June 2002, p. 5985–5992. 10.1128/JVI.76.12.5985-5992.2002 https://jvi.asm.org/content/76/12/5985
- Sprygin A, Babin Y, Pestova Y, Kononova S, Wallace DB, Van Schalkwyk A, et al. (2018) Analysis and insights into recombination signals in lumpy skin disease virus recovered in the field. PLoS ONE 13(12): e0207480. https://doi.org/ 10.1371/journal.pone.0207480
- Liitsola, K., Holm K., Bobkov, A., Pokrovsky, V., Smolskaya,T., Leinikki,P., Osmanov,S. and Salminen,M. (2000) An AB recombinant and its parental HIV type 1 strains in the area of the former Soviet Union: low requirements for sequence identity in recombination. UNAIDS Virus Isolation Network. AIDS Res. Hum. Retroviruses, 16, 1047–1053.
- Smith, D. B., Bukh, J., Kuiken, C., Muerhoff, A. S., Rice, C. M., Stapleton, J. T., & Simmonds, P. (2014). Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology, 59(1), 318–327. https://doi.org/10.1002/hep.26744
- Jiang,X., Espul,C., Zhong,W.M., Cuello,H. and Matson,D.O. (1999) Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol., 144, 2377–2387