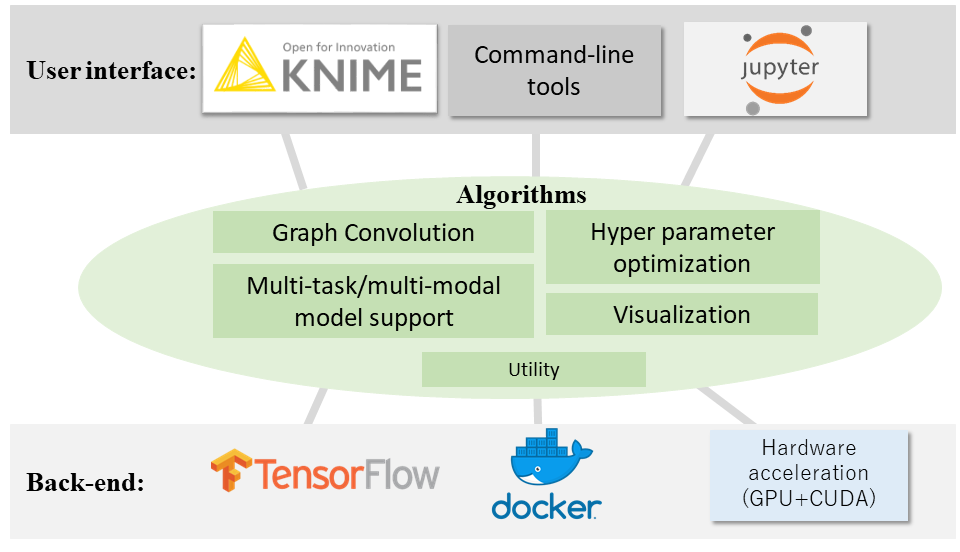
A setup script is under construction. Now, you have to execute the python codes directly.
- python: >3.6
- tensorflow: >1.12 (partially suporting TensorFlow2 by the compatible mode, but does NOT guarantee working on TensorFlow2)
- joblib
- numpy
- scipy
- scikit-learn: >0.21
- matplotlib
To install additional modules:
- Visualizer: please see gcnvisualizer/README.md
- KNIME: please see KNIME/README.md
- Jupyter Notebook: please see Notebook/README.md
- Docker: https://hub.docker.com/r/clinfo/kgcn
This is a TensorFlow implementation of Graph Convolutional Networks for the task of classification of graphs.
Our implementation of Graph convolutional layers consulted the following paper:
Thomas N. Kipf, Max Welling, Semi-Supervised Classification with Graph Convolutional Networks (ICLR 2017)
For training with a dataset, example_jbl/synthetic.jbl, by using a neural network defined in example_model/model.py
kgcn train --config example_config/sample.jsonwhere sample.json is a config file.
For testing and inferrence
kgcn infer --config example_config/sample.json --model model/model.sample.last.ckptwhere model/model.sample.last.ckpt is a trained model file.
Our sample dataset file (example.jbl) is created by the following command:
python example_script/make_example.pyWhen you create your own dataset, you can refer make_sample.py. This script converts adjacency matrices (example_data/adj.txt), features (example_data/feature.txt), and labels (example_data/label.txt) into the dataset file (example_jbl/sample.jbl)
For example, in training phases, you can specify a dataset as follows:
kgcn train --config example_config/sample.json --dataset example_jbl/sample.jblYou can specify a configuration file (example_config/sample.json) as follows:
kgcn train --config example_config/sample.jsonkgcn has three commands: train/infer/train_cv. You can specify a command as follows:
kgcn <command> --config example_config/sample.json-
train command: The script trains a model and saves it.
-
infer command: The script estimates labels of test data using the loaded model.
-
train_cv command: The command simplifies cross-validation routines including training stages and estimation(evaluation) stages. Once you execute this command, cross-validation is performed by running a seriese of training and estimation programs.
Dataset file: To use your own data, you have to create a dictionary with the following data format and compress it as a joblib dump file.
Another PyTorch-based library is also available:
In the current version of kMoL, it is not completely compatible with kGCN, but we are developing the kMoL library with the aim of compatibility.
@article{Kojima2020,
author = "Ryosuke Kojima and Shoichi Ishida and Masateru Ohta and Hiroaki Iwata and Teruki Honma and Yasushi Okuno",
title = "{kGCN: a graph-based deep learning framework for chemical structures}",
year = "2020",
month = "5",
journal = "Journal of Cheminformatics",
volume = "12",
number = "1",
url = "https://jcheminf.biomedcentral.com/articles/10.1186/s13321-020-00435-6",
doi = "10.1186/s13321-020-00435-6"
}
.
├── active_learning/ :
├── data_generator/ :
│ ├── synth_generator.py : random graph
│ └── synth_generator_ring.py : random graph with ring
├── docs/ : a set of documents
├── example_config/ : examples of config files
├── example_data/ : examples of adj. files, label files, etc.
├── example_jbl/ : examples of jbl. files
├── example_model/ : examples of model files
├── example_param/ : examples of parameter domain files
├── example_script/ : scripts for the examples
├── gcn_modules/ :
├── gcnvisualizer/ : kgcn visualization modules
├── graph_kernel/ : graph kernel SVM
├── hooks/ :
├── kgcn :
│ ├── legacy : duplicated scripts
│ ├── preprocessing/ : scripts for dataset preparaiton for kgcn
│ ├── core.py : a main program files for the GCN model
│ ├── data_util.py : utilities for data handling
│ ├── default_model.py :
│ ├── error_checker.py : error checker
│ ├── feed.py : functions to build feed dictionaries
│ ├── feed_index.py : functions to build feed dictionaries (index base)
│ ├── layers.py : GCN-related layers
│ ├── make_plots.py : functions to plot graphs
│ └── visualization.py : functions to visualize trained models
├── kgcn_tf2 :
├── kgcn_torch :
├── KNIME/ :
├── logs/ : output directory for exmaples
├── model/ : output directory for exmaples
├── Notebook/ : examples of jupyter notebook
├── result/ : output directory for exmaples
├── sample_kg/ :
├── sample_chem/ :
├── sample_nx/ :
├── script : utility sctipts
│ ├── make_dataset.py :
│ ├── plot_graph.py :
│ ├── show_graph.py :
│ └── show_label_balance.py :
├── script_cv : scripts for parallel cross validation
│ ├── 01make_dataset.sh :
│ ├── 02run_fold.sh :
│ └── make_cross_validation_dataset.py :
├── visualization/ : output directory for exmaples
├── Dockerfile :
├── gcn.py : the main engin of this project
├── gcn_gen.py : an engin for generative models
├── gcn_pair.py : an engin for ranking models
├── LICENSE : LICENSE file
├── model_functions.py :
├── opt_hyperparam.py :an engin for optimization of hyper parameters
├── README.md : this file
├── setup.py :
└── task_sparse_gcn.py :
| command | python file | description |
|---|---|---|
| kgcn | gcn.py | a main command of kGCN for learning and prediction |
| kgcn-gen | gcn_gen.py | a command for generative models (learning, reconstruction, and generation) |
| kgcn-sparse | task_sparse_gcn.py | a command for on-the-fly learning and prediction using tfrecords |
| command | python file | description |
|---|---|---|
| kgcn-chem | kgcn/preprocessing/chem.py | a command to preprocess chemical compounds and protein data |
| kgcn-kg | kgcn/preprocessing/kg.py | a command to preprocess knowledge graph data |
| kgcn-cv-splitter | script_cv/cv_splitter | a command to split a dataset file(.jbl) for cross-validation (especially for parallel execution) |
| kgcn-join | kgcn/data_join | a command to join dataset files(.jbl) |
| command | python file | description |
|---|---|---|
| kgcn-opt | opt_hyperparam.py | a command for hyper parameter optimization using optuna library |
| gcnv | gcnvisualize/ | a command for visualization (see https://github.com/clinfo/kGCN/tree/master/gcnvisualizer ) |
We provide additional example using synthetic data to discriminate 5-node rings and 6-node rings. The following command generates synthetic data as text formats:
python data_generator/synth_generator_ring.pyThe following command generates .jbl from text:
python example_script/make_synth.pyThe following command carries out cross-validation:
kgcn train_cv --config example_config/synth.jsonAccuracy and the other scores are stored in:
result/synth_cv_result.json
More information is stored in:
result/synth_info.json
We prepared additional samples for multimodal and multitask learning. You can specify a configuration file (sample_multimodal_config.json/sample_multitask_config.json) as follows:
kgcn --config example_config/multimodal.json trainFor multimodal, symbolic sequences and graph data are used as the inputs of a neural network. This configuration file specifies the program of model as "model_multimodal.py", which includes definition of neural networks for graphs, sequences, and combining them. Please reffer to sample/seq.txt and a coverting program (make_example.py) to prepare sequence data,
kgcn --config example_config/multitask.json trainIn this sample, "multitask" means that multiple labels are allowed for one graph. This configuration file specifies the program of model as "model_multitask.py", which includes definition of a loss function for multiple labels. Please reffer to sample_data/multi_label.txt and a coverting program (make_sample.py) to prepare multi labeled data,
python gcn_gen.py --config example_config/vae.json traingcn_gen.py is an alternative gcn.py for generative models. example_config/vae.json is a setting for VAE (Variational Auto-encoder) that is implemented in example_model/model_vae.py
First, prepare .tfrecords files in a dataset folder.
The files that are named '[train, eval, test].tfrecords' are used for training, eval, test.
You can have multiple files for training, etc. Alternatively, you can just have one file that contains multiple examples for training.
The format of serialized data in .tfrecords:
features = {
'label': tf.io.FixedLenFeature([label_length], tf.float32),
'mask_label': tf.io.FixedLenFeature([label_length], tf.float32),
'adj_row': tf.io.VarLenFeature(tf.int64),
'adj_column': tf.io.VarLenFeature(tf.int64),
'adj_values': tf.io.VarLenFeature(tf.float32),
'adj_elem_len': tf.io.FixedLenFeature([1], tf.int64),
'adj_degrees': tf.io.VarLenFeature(tf.int64),
'feature_row': tf.io.VarLenFeature(tf.int64),
'feature_column': tf.io.VarLenFeature(tf.int64),
'feature_values': tf.io.VarLenFeature(tf.float32),
'feature_elem_len': tf.io.FixedLenFeature([1], tf.int64),
'size': tf.io.FixedLenFeature([2], tf.int64)
}
Then, run following command.
python task_sparse_gcn.py --dataset your_dataset --other_flagskgcn-opt --config ./example_config/opt_param.json --domain ./example_param/domain.jsonkgcn-opt (opt_hyperparam.py) is a command for hyperparameter optimization using GPyOpt library (https://github.com/SheffieldML/GPyOpt), a Bayesian optimization libraly. ./example_config/opt_param.json is a config file to use gcn.py ./example_param/domain.json is a domain file to define hyperparameters and their search spaces. The format of this file follows "domain" of GPyOpt. For more information for this json file, see the GPyOpt document(http://nbviewer.jupyter.org/github/SheffieldML/GPyOpt/blob/devel/manual/index.ipynb ).
Depending on your environment, it might be necessary to change line 9 (opt_cmd) of opt_hyperparam.py
When you want to change and add hyperparameters, please change domain.json and model file. An example of model file is example_model/opt_param.py in which a hyperparameter is num_gcn_layer.
This edition of kGCN is for evaluation, learning, and non-profit
academic research purposes only, and a license is needed for any other uses.
Please send requests on license or questions to kojima.ryosuke.8e@kyoto-u.ac.jp.