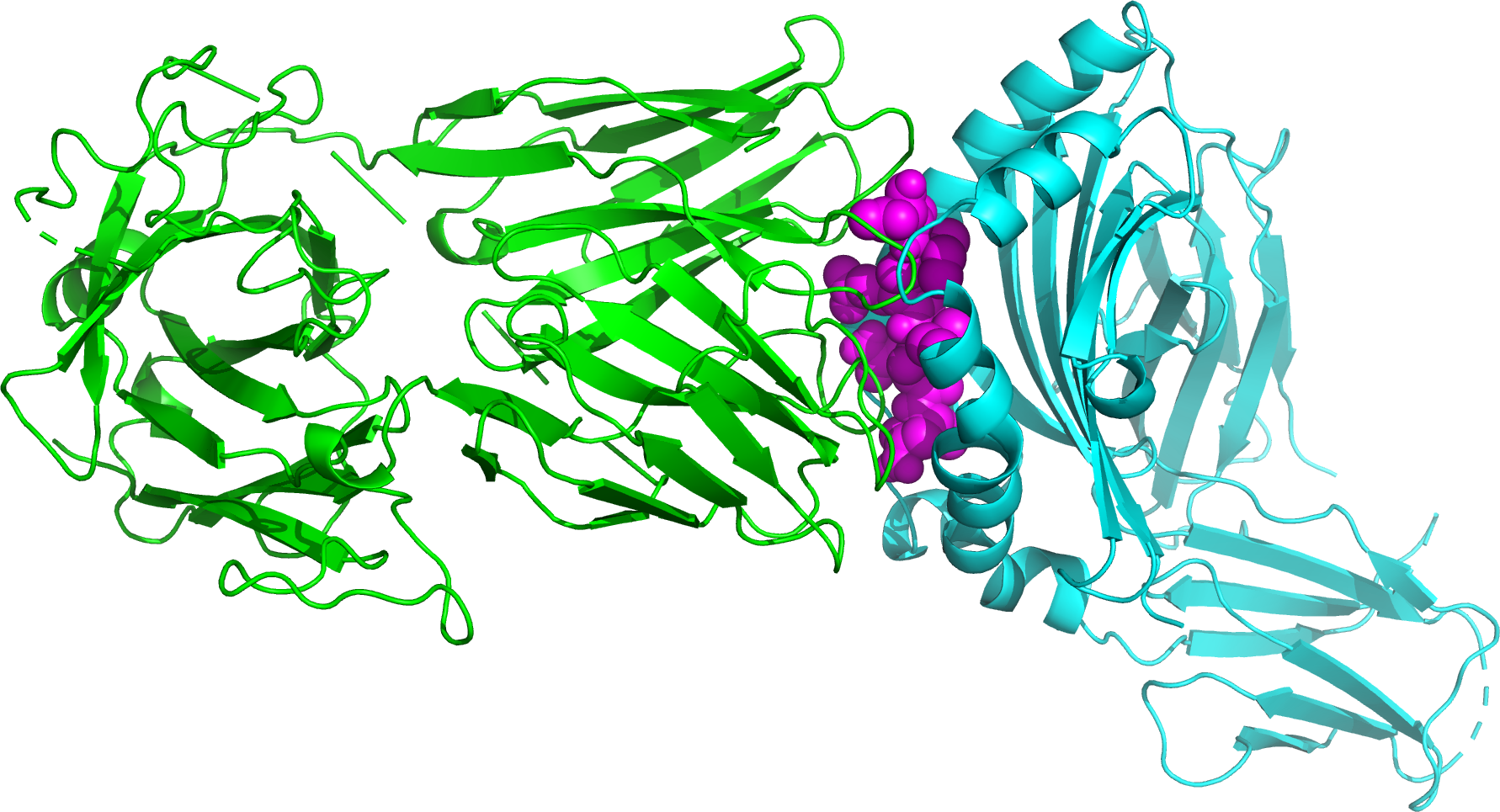
Genetic variations of Plasmodium falciparum circumsporozoite protein and the impact on interactions with human immunoproteins and malaria vaccine efficacy
Cheikh Cambel Dieng, Colby T. Ford, Anita Lerch, Dickson Doniou, Jennifer Huynh, Kovidh Vegesna,
Jun-tao Guo, Daniel Janies, Liwang Cui, Linda Amoah, Yaw Afrane, and Eugenia Lo
Published in Infection, Genetics and Evolution:
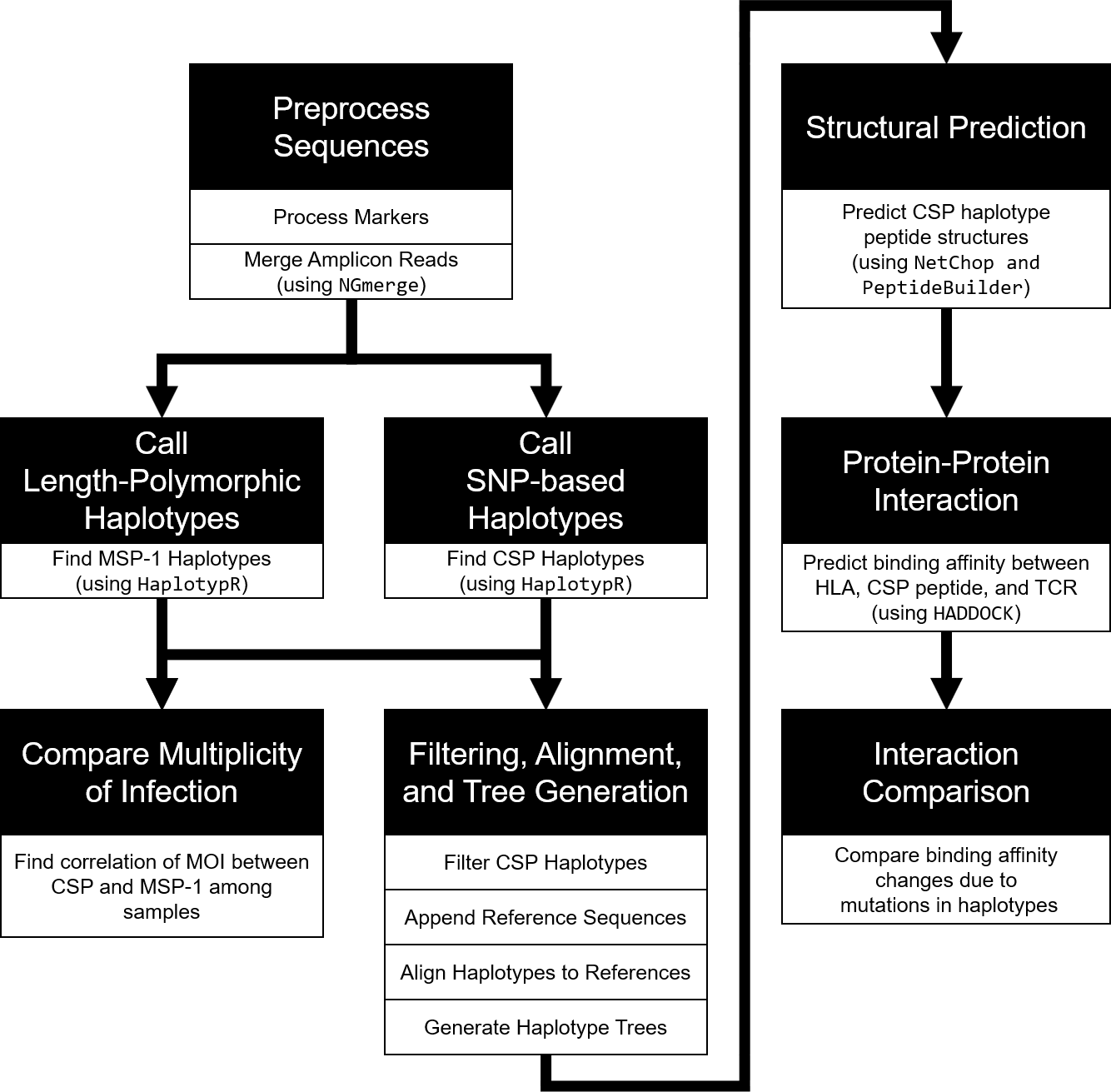
In October 2021, the world's first malaria vaccine RTS,S was endorsed by WHO for broad use in children, despite its low efficacy. This study examined polyclonal infections and the associations of parasite genetic variations with binding affinity to human leukocyte antigen (HLA). Multiplicity of infection was determined by amplicon deep sequencing of PfMSP1. Genetic variations in PfCSP were examined across 88 samples from Ghana and analyzed together with 1655 PfCSP sequences from other African and non-African isolates. Binding interactions of PfCSP peptide variants and HLA were predicted using NetChop and HADDOCK. High polyclonality was detected among infections, with each infection harboring multiple non-3D7 PfCSP variants. Twenty-seven PfCSP haplotypes were detected in the Ghanaian samples, and they broadly represented PfCSP diversity across Africa. The number of genetic differences between 3D7 and non-3D7 PfCSP variants does not influence binding to HLA. However, CSP peptide length after proteolytic degradation significantly affects its molecular weight and binding affinity to HLA. Despite the high diversity of HLA, the majority of the HLAI and II alleles interacted/bound with all Ghana CSP peptides. Multiple non-3D7 strains among P. falciparum infections could impact the effectiveness of RTS,S. Longer peptides of the Th2R/Th3R CSP regions should be considered in future versions of RTS,S.
- CSP_sequence_analysis/
- Final List of CSP Haplotypes (SNP-based): finalTab_csp.csv
- Haplotype Creation Script: MalariaHaplotyping_NGmerge_script.R
- Haplotype Network Resources: haplotype_network/
- Protein Modeling Resources: protein/
- HADDOCK Docking Results: HADDOCK_Results.xlsx
- CD4+/Th2R Protein Docking Files: CD4_Th2R/
- CD8+/Th3R Protein Docking Files: CD8_Th3R/
- MSP1_sequence_analysis/
- Final List of MSP1 Haplotypes: finalTab_msp.csv
- Haplotype Creation Script: MalariaHaplotyping_NGmerge_script.R
- CSP-MSP1_Comparison/
- Multiplicity of Infection by Sample: MSP-CSP_mMOI_Comparison.xlsx
All Amplicon-seq paired-end FASTQ files of CSP and MSP-1 genes are available on NCBI SRA: https://www.ncbi.nlm.nih.gov/sra/PRJNA783000
- HaplotypR pacakge: https://github.com/lerch-a/HaplotypR
- NetChop 3.1 Server: http://www.cbs.dtu.dk/services/NetChop/
- PeptideBuilder package: https://github.com/clauswilke/peptidebuilder
- HADDOCK Server: https://wenmr.science.uu.nl/haddock2.4/
- PyMol: https://pymol.org/
- PDB Structures: