NOTICE: README IS OUTDATED - NOW A PART OF THIS PROJECT: https://github.com/danjonpeterson/print-my-brain.git
Do you have an MRI of your Brain?
Do you want to get an .stl file of your brain so that you can make 3D print of it?
All you need is Docker.
This project describes the creation of a docker image that can take an MRI scan and output an .stl file suitable for 3d printing.
It also generates a rotating .gif of that .stl file so that you can see if it's worth printing.
For this to work, you need a high-quality, high-resolution T1-weighted image - sometimes called an MPRAGE.
The docker image itself is pretty large (>8GB) and may take a while to build. The processing itself can take more than a full day to run.
- install docker
- clone this repository
git clone https://github.com/danjonpeterson/brain_printer.git
- build the image from the Dockerfile, call it brain_printer
cd brain_printer
docker build -t brain_printer .
- put your brain image (in nifti or nifti_gz format) in an empty directory, say print_my_brain/.
If your image is is DICOM format, take a look at https://www.nitrc.org/projects/dcm2nii/
mkdir print_my_brain/
cp my_brain.nii.gz print_my_brain/
- run the docker image, mounting your newly created directory in /data
docker run -ti --rm -v /full/path/to/print_my_brain/:/data brain_printer
- from the docker image, run the script: run.sh
/brain_printer/run.sh
If everything works well, in your new directory you should see these files:
rh.stl - file for 3d printing the right hemisphere
lh.stl - likewise, for the left
rh.gif - an animated gif showing you the model for the right hemisphere
lh.gif - likewise, for the left
001/ - a freesurfer processing directory that has lots of information about the brain
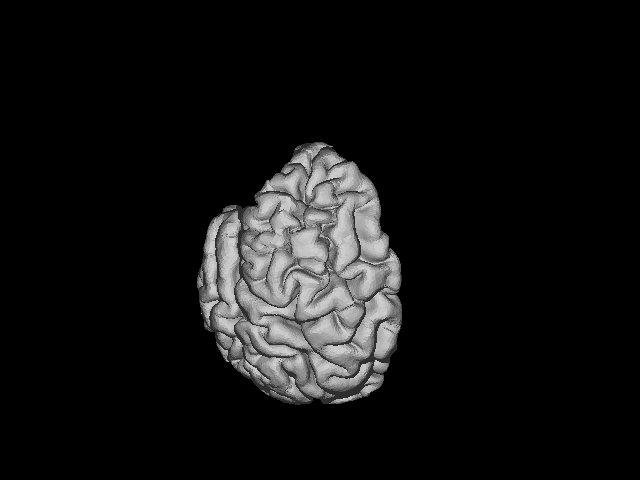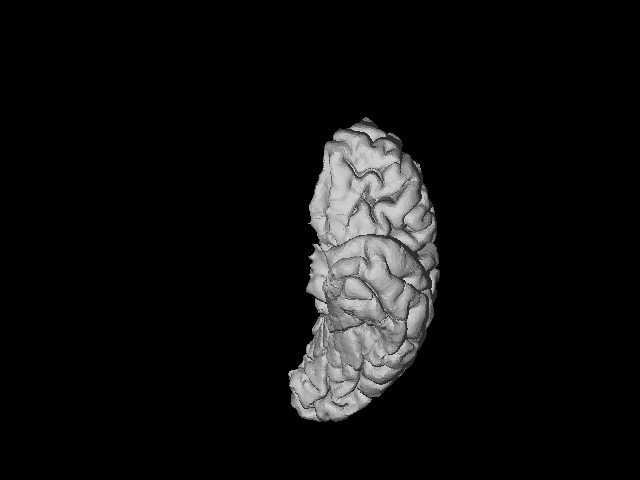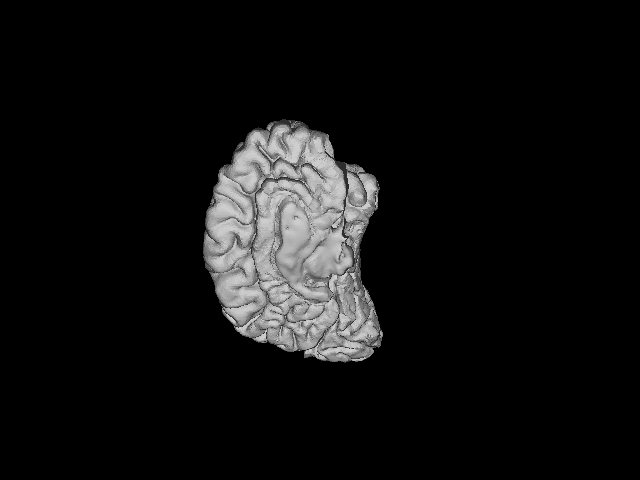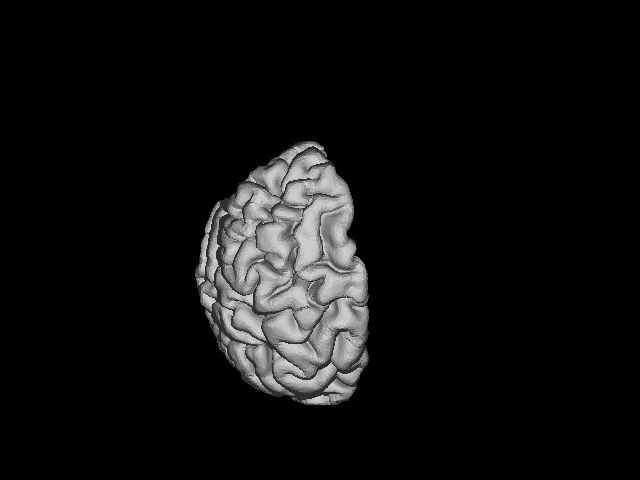
The .gifs look like this:
LEFT HEMISPHERE:
RIGHT HEMISPHERE:
This was created as part of neurohackweek2016. It uses freesurfer (https://surfer.nmr.mgh.harvard.edu/) to extract the cortical surface. You really should get your own license file https://surfer.nmr.mgh.harvard.edu/fswiki/DownloadAndInstall#License and put it in your copy of the docker image.
Also loaded in the docker image is:
stl2pov https://github.com/timschmidt/stl2pov
povray http://povray.org/
imagemagick www.imagemagick.org
Thanks to Valentia Staneva, Matteo Visconti, Chris Madan and everyone else at #NHW16
Writeup here:
https://doi.org/10.3897/rio.3.e13394
- fully test freesurfer the whole way through (not done for this ~4 day project - it was faked by copying files over from a completed freesurfer run)
- dicom support
- smooth/decimate mesh
- make standalone html file to contain the .gifs
Freesurfer can be tempermental, depending on the quality of the input image. There is an active and searchable listerv/forum on the Freesurfer website (http://www.mail-archive.com/freesurfer@nmr.mgh.harvard.edu/).
One user had some conneciton issues installing some of the packages during the container build. This was resolved by editing the docker config as described here: https://stackoverflow.com/questions/24991136/docker-build-could-not-resolve-archive-ubuntu-com-apt-get-fails-to-install-a?rq=1