#Abstracting Real World Data from Oncology Notes
In this collection, we use John Snow Labs’ Spark NLP for Healthcare, the most widely-used NLP library in the healthcare and life science industries, to extract, classify and structure clinical and biomedical text data with state-of-the-art accuracy at scale. For this solution we used the [MT ONCOLOGY NOTES]((https://www.mtsamplereports.com/) dataset. It offers resources primarily in the form of transcribed sample medical reports across medical specialties and common medical transcription words/phrases encountered in specific sections that form part of a medical report – sections such as physical examination or PE, review of systems or ROS, laboratory data and mental status exam, among others.
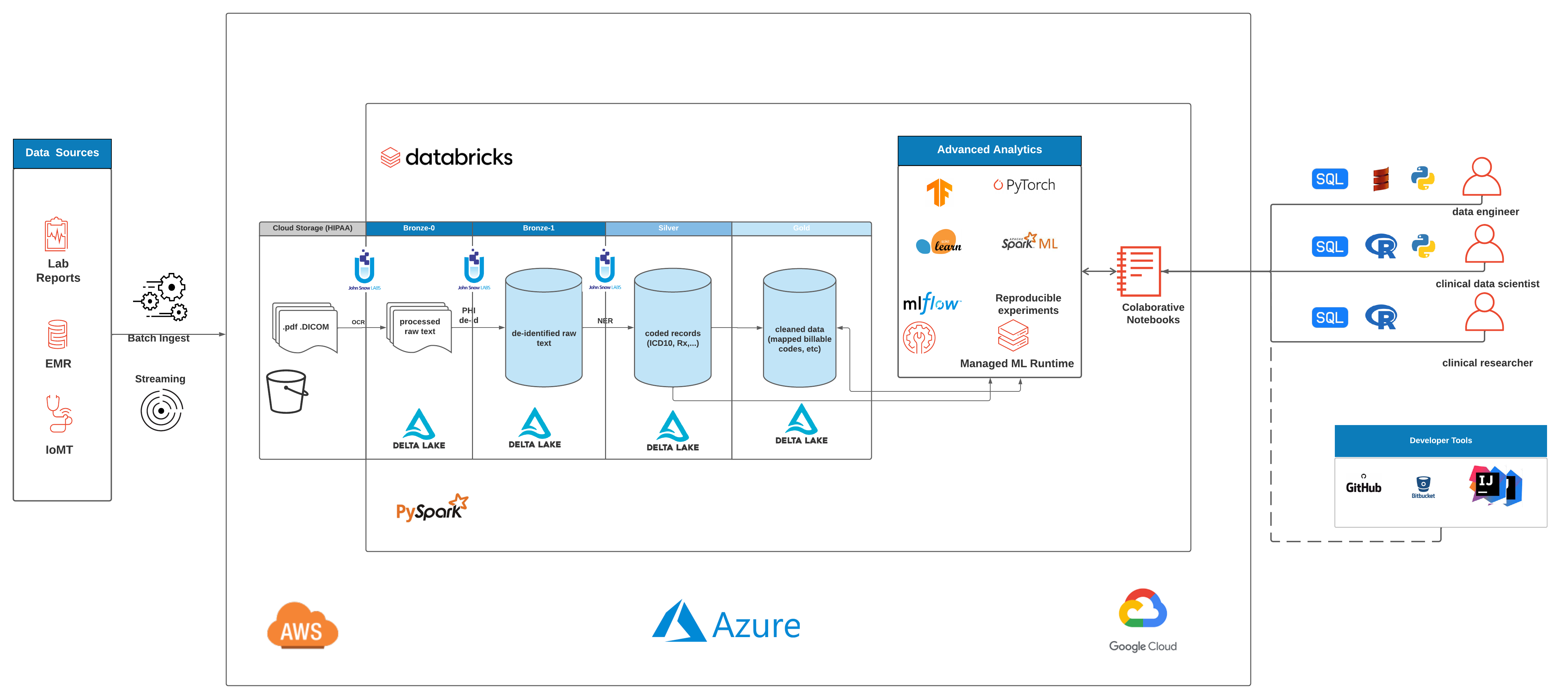
We chose 50 de-identified oncology reports from the MT Oncology notes dataset as the source of the unstructured text and landed the raw text data into the Delta Lake bronze layer. For demonstration purposes, we limited the number of samples to 50, but the framework presented in this solution accelerator can be scaled to accommodate millions of clinical notes and text files.
The first step in our accelerator is to extract variables using various models for Named-Entity Recognition (NER). To do that, we first set up our NLP pipeline, which contains annotators such as documentAssembler and sentenceDetector and tokenizer that are trained specifically for healthcare-related NER.
We then create dataframes of extracted entities and land the tables in Delta where can be accessed for interactive analysis or dashboarding using databricks SQL.
MT ONCOLOGY NOTES comprises of millions of ehr records of patients. It contains semi-structured data like demographics, insurance details, and a lot more, but most importantly, it also contains free-text data like real encounters and notes.
Here we show how to use Spark NLP's existing models to process raw text and extract highly specialized cancer information that can be used for various downstream use cases, including:
- Staff demand analysis according to specialties.
- Preparing reimbursement-ready data with billable codes.
- Analysis of risk factors of patients and symptoms.
- Analysis of cancer disease and symptoms.
- Drug usage analysis for inventory management.
- Preparing timeline of procedures.
- Relations between internal body part and procedures.
- Analysis of procedures used on oncological events.
- Checking assertion status of oncological findings.
There are three notebooks in this package:
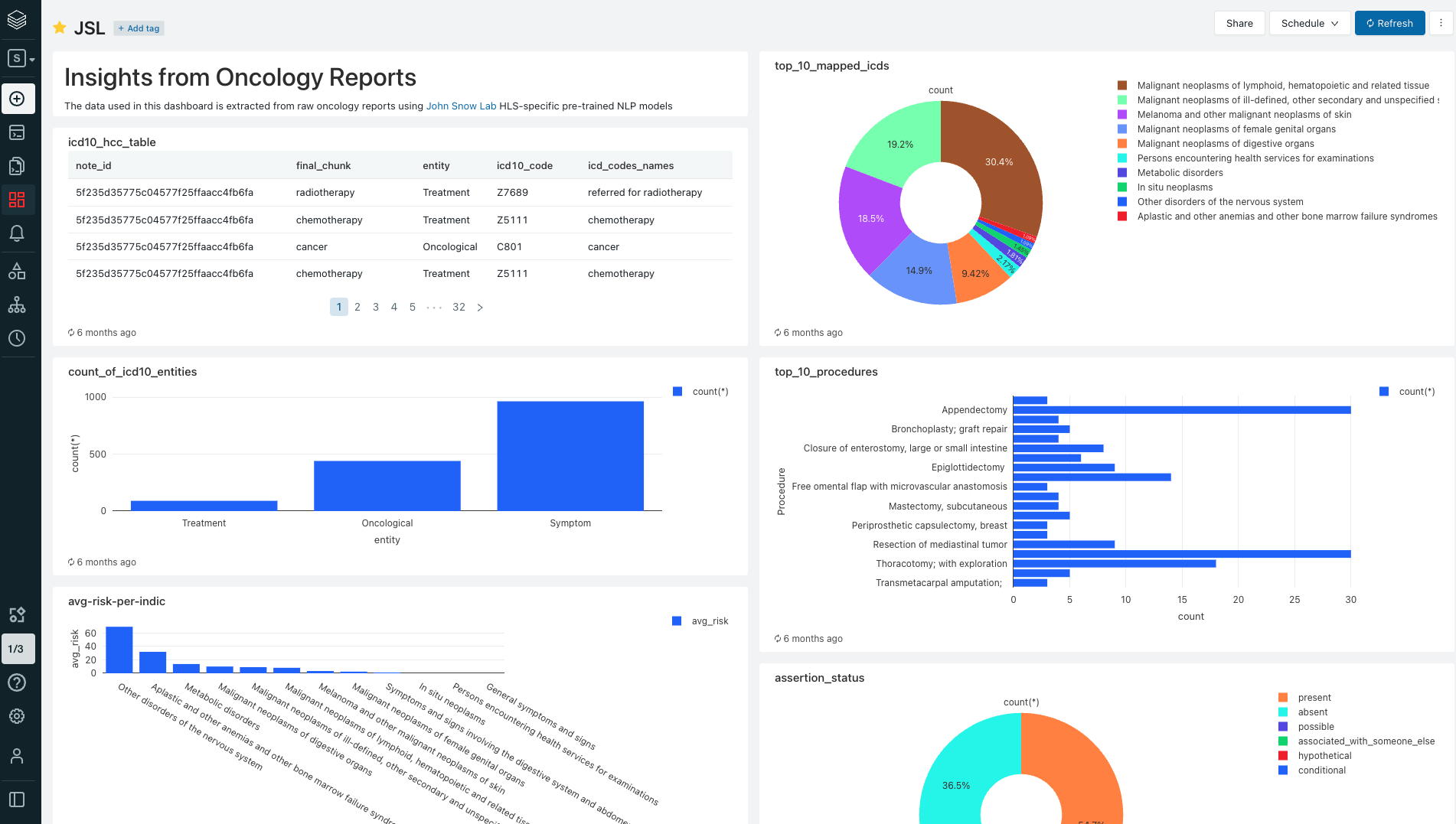
config: Notebook for configuring the environmententity-extraction: Extract drugs, oncological entities, assertion status and relationships and writes the data into Delta lake.oncology-analytics: Interactive analysis of the data.
Copyright / License info of the notebook. Copyright [2021] the Notebook Authors. The source in this notebook is provided subject to the Apache 2.0 License. All included or referenced third party libraries are subject to the licenses set forth below.
| Author |
|---|
| Databricks Inc. |
| John Snow Labs Inc. |
Databricks Inc. (“Databricks”) does not dispense medical, diagnosis, or treatment advice. This Solution Accelerator (“tool”) is for informational purposes only and may not be used as a substitute for professional medical advice, treatment, or diagnosis. This tool may not be used within Databricks to process Protected Health Information (“PHI”) as defined in the Health Insurance Portability and Accountability Act of 1996, unless you have executed with Databricks a contract that allows for processing PHI, an accompanying Business Associate Agreement (BAA), and are running this notebook within a HIPAA Account. Please note that if you run this notebook within Azure Databricks, your contract with Microsoft applies.
The job configuration is written in the RUNME notebook in json format. The cost associated with running the accelerator is the user's responsibility.
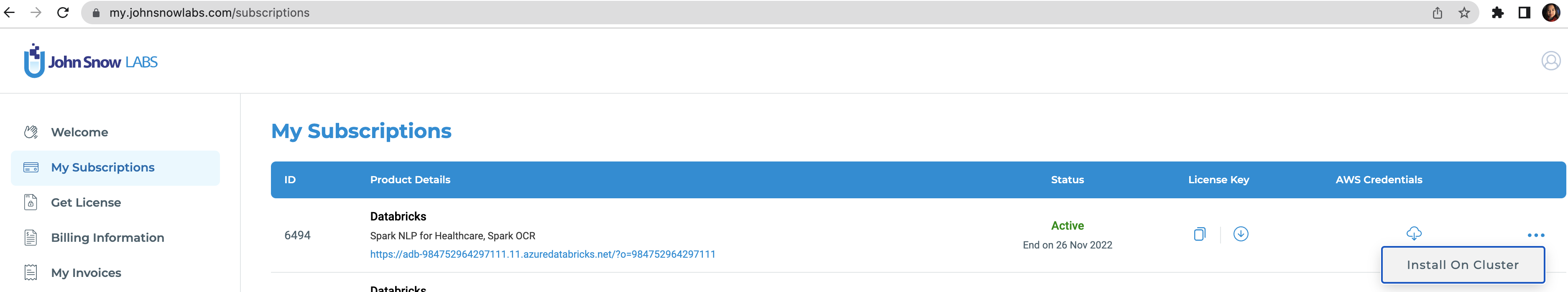
To run this accelerator, set up JSL Partner Connect AWS, Azure and navigate to My Subscriptions tab. Make sure you have a valid subscription for the workspace you clone this repo into, then install on cluster as shown in the screenshot below, with the default options. You will receive an email from JSL when the installation completes.
Once the JSL installation completes successfully, clone this repo into a Databricks workspace. Attach the RUNME notebook to any cluster running a DBR 11.0 or later runtime, and execute the notebook via Run-All. A multi-step-job describing the accelerator pipeline will be created, and the link will be provided. Execute the multi-step-job to see how the pipeline runs.