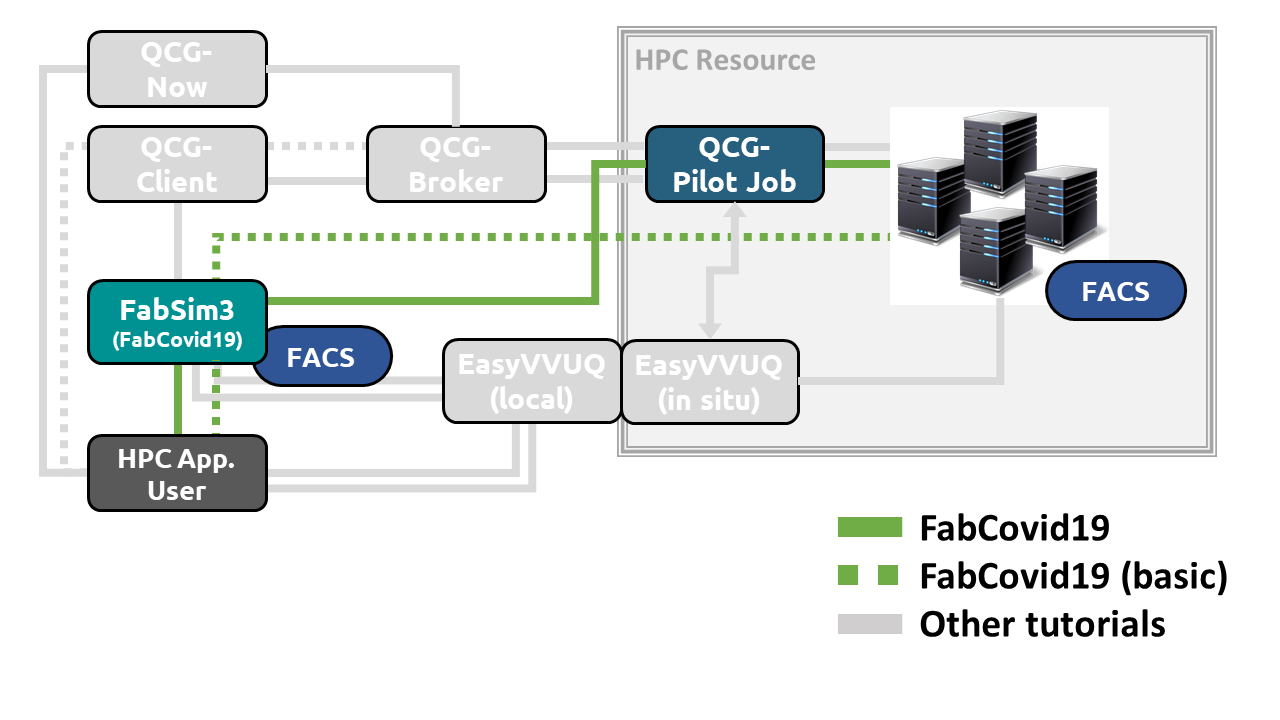
FabCovid19 is a FabSim3 plugin for Flu And Coronavirus Simulator (FACS).
A step-by-step guide for Execution of FACS Simulation at sub-national level using local and HPC resources
This is a shortened tutorial. Full documentation for FabCovid19 is available at facs.readthedocs.io.
In this tutorial, you will get step-by-step guidance on the usage of FabCovid19 components to simulate FACS simulations within a local and HPC execution environment. In this tutorial you will learn about the following FabCovid19 software components and how these components are used in COVID-19 prediction application as shown in the Tube Map below:
- FabSim3 - an automation toolkit that features an integrated test infrastructure and a flexible plugin system.
- FACS - an agent based simulation framework that models the viral spread at the sub-national level, incorporating geospatial data sources to extract buildings and residential areas within a predefined region.
- QCG Pilot Job - a Pilot Job system that allows to execute many subordinate jobs in a single scheduling system allocation.
- Infectious Disease simulations
- Dependencies
- Installation of required software packages
- FabSim3 Configuration
- Execution
- Acknowledgements
FabCovid19 is a FACS toolkit plugin for infectious disease simulation which automates complex simulation workflows. In this tutorial, we demonstrate different types of FACS simulations. We explain how you can do create an agent-based disease transmission model of a specific infectious disease e.g., COVID-19 and forecast its spread over space and time. This tutorial demonstrates the model construction and steps to perform a set of runs based on different lockdown scenarios, and visualize the spread of infections across space and time with confidence intervals.
Suggestion: To make path configuration easier, we recommend creating a dedicated work directory where you install both FabSim3 and FACS. This will help ensure straightforward path realization and make it easier to manage related path dependencies.
- FabSim3
To install the Fabsim3 automation toolkit, see the installation documentation or following the installation, go to
(FabSim3 Home)directory and executepython configure_fabsim.py. This script is designed to quickly configure FabSim3. - FACS: Flu And Coronavirus Simulator
To install FACS in your working directory, simply type:
git clone https://github.com/djgroen/facs.gitTo install FabCovid19, simply go to the FabSim3 directory and type:
fabsim localhost install_plugin:FabCovid19After cloning the required dependencies, a few configuration steps are needed to run FACS using FabCovid19 in FabSim3.
To execute FACS in FabSim3 using FabCovid19 plugin, FabSim3 needs the full path to your FACS installation. You can specify this path in two configuration files: machines_user.yml and machines_FabCovid19_user.yml.
- Navigate to
(FabSim3 Home)/deploy: - Open
machines_user.yml. Ifmachines_user.ymlis not present, copymachines_user_example.ymltomachines_user.yml. - Under the
default:section, add the following line:
facs_location: "(FACS PATH)"Note: Replace (FACS PATH) with the full path to your FACS installation, such as /home/fabuser/facs.
Alternatively, you can add the FACS path directly within FabCovid19:
- In
(FabSim3 Home)/plugins/FabCovid19, copymachines_FabCovid19_user_example.ymltomachines_FabCovid19_user.yml. - Open
machines_FabCovid19_user.ymland locatefacs_location: "<..>", and Replace<..>with the full path to your FACS installation, such as:
facs_location: "/home/fabuser/facs"If you would like to customize other FabSim3 or FabCovid19 parameters, you can modify them within machines_user.yml and machines_FabCovid19_user.yml as needed.
Note: If you are installing on MacOS, remember that path conventions may differ (e.g., replace /home with /Users in paths like /Users/yourusername/facs).
The current supported arguments for running FACS are listed below. These allow users to customize simulation parameters such as location, starting infections, disease configuration, and more:
- --location: Sets the location for the simulation (e.g., --location=test).
- --generic_outfile: Specifies the name of the output file (default: output.txt).
- --output_dir: Defines the directory where output files are saved (default: .).
- --measures: Specifies intervention measures for the simulation (left blank if no measures).
- --data_dir: Directory for COVID-related data files (e.g., --data_dir=covid_data).
- --starting_infections: Initial number of infections at the start of the simulation (e.g., --starting_infections=1).
- --start_date: Start date of the simulation in dd/mm/yyyy format (e.g., --start_date=1/3/2020).
- --simulation_period: Duration of the simulation in days. Use -1 for an indefinite period.
- --household_size: Average household size for the simulation population (e.g., --household_size=2.6).
- --disease_yml: Specifies the YAML configuration file for disease parameters (e.g., --disease_yml=disease_covid19).
python run.py --location=test --output_dir=. --measures=measures_uk --data_dir=covid_data --starting_infections=1 --start_date=1/3/2020 --simulation_period=1 --household_size=2.6 --disease_yml=disease_covid19This example sets up a simulation with 10 initial infections, running for one day, with data sourced from covid_data and using the disease_covid19.yml configuration file. The output is generated in form of covid_out_*.csv (e.g., covid_out_deaths_0.csv) for individual parameters or as test-measures_uk.csv in facs home directory.
The FabCovid19 command structure is fabsim <local/remote machine> <function>:<config-file(s)>,<arg1,arg2,arg3,...>.
We can construct a simple command with two arguments as follow:
fabsim localhost covid19:config=test,cores=1,starting_infections=1FabSim3 creates a bash executable in /path/to/FabSim3/localhost_exe sub-directories as define in machines_user.yml or machines_FabCovid19_user.yml and executes the simulation on localhost.
Note: Please see the printouts in terminal for more runtime information.
After the simulation is completed, we can fetch the simulation outputs by typing:
fabsim localhost fetch_resultsThe output is fetched and stored in /path/to/FabSim3/results directory.
FACS team have created more than 20 locations (config files) as part of collaboration in STAMINA project. These locations are brent, ealing, harrow, hillingdon and more. Please see FabCovid19/config_files directory.
In FACS, a simulation of a geographical location can be defined as a task. A FabSim3 job is either simulation of a task or many tasks. In the example below we submit a job containing one task.
fabsim localhost covid19:config=harrow,measures=measures_uk,cores=1,starting_infections=1In ensemble more, we will submit a job containing multiple tasks or many tasks, each executed one or more times. In this example, we replicate test twice replicas=2 and execute it twice.
fabsim localhost covid19_ensemble:configs='test',cores=1,starting_infections=1,replicas=2Or we can run multiple tasks, multiple times:
fabsim localhost covid19_ensemble:configs='test;brent',cores=1,starting_infections=1,replicas=2Up to now, we executed our FACS jobs on localhost using FabCovid19 plugin in FabSim3, however, we can submit our jobs to other machines to be executed remotely. FabSim3 provides several remote machines configurations such as ARCHER2.
The examples mentioned above can be executed remotely by replacing the machine section of the command:
fabsim archer2 covid19:config=test,cores=1,starting_infections=1Note: Before executing, please ensure the remote machines configuration is added to the machines_user.yaml or machines_FabCovid19_user.yml. An example of the configuration is added below:
archer2:
username: <your-username>
manual_ssh: true
remote: archer2
budget: <your-budget>
project: <your-project>
job_wall_time: '0-00:10:00'
job_dispatch: 'sbatch'
partition_name: "standard"
qos_name: "short"FabSim3 currently supports QCG-PilotJob and Radical-Pilot.
All ensemble jobs can be executed in pilot mode by added arguments PJ and PJ_TYPE:
fabsim archer2 covid19_ensemble:configs='test',cores=16,starting_infections=1,PJ=True,PJ_TYPE=QCGUse PJ_TYPE=RP for Radical-Pilot job execution.
This work was supported by the HiDALGO, VECMA and STAMINA projects, which has received funding from the European Union Horizon 2020 research and innovation programme under grant agreement No 824115, 800925 and 883441.
If you encountered any problem during the installation, configuration and execution, please raise a GitHub issue.