单细胞测序的分析模型汇总和学习笔记
Model zoo and study notes for single cell data analysis.
- Single-cell best practices (by Theislab) https://www.sc-best-practices.org/
- Analysis of single cell RNA-seq data https://www.singlecellcourse.org/
- Orchestrating Single-Cell Analysis with Bioconductor https://bioconductor.org/books/release/OSCA/
- Best Practices for Spatial Transcriptomics Analysis with Bioconductor https://lmweber.org/BestPracticesST/
- Best practices for single-cell analysis across modalities (2023) [paper]
- Current best practices in single‐cell RNA‐seq analysis: a tutorial (2019) [paper]
-
Human Cell Atlas [Home] [Data Portal] [paper] [pdf]
- Human Developmental Cell Atlas [UK Team] [Sweden Team] [Roadmap]
-
CZ CELLxGENE [Home] [CellGuide] [Census package] [Census R package]
-
Dixit et al, 2016. [Article] [GEO] [Harvard Dataverse] [Regev Lab] [pdf]
-
Adamson et al, 2016. [Article] [GEO] [Harvard Dataverse] [Weissman Lab] [pdf]
- Direction of perturbations: CRISPRi (gene knockdown)
- Number of perturbed genes: one-gene perturbations
- Cell type: K562
-
Norman et al, 2019. [Article] [BioRxiv] [GEO] [Harvard Dataverse] [Weissman Lab] [pdf]
- Direction of perturbations: CRISPRa (gene activation)
- Number of perturbed genes: one-gene perturbations and two-gene perturbations
- Cell type: K562
-
Papalexi et al, 2021. [Article] [BioRxiv] [GEO] [Satija Lab] [Vignette]
-
Replogle et al, 2022. [Article] [Data Portal] [GitHub1] [GitHub2] [Weissman Lab] [pdf]
- scPerturb.org Single Cell Perturbation Datasets [Home] [GitHub] [Sander Lab]
- limma (2003). [Home] [Article] [Bioconductor] Linear Models for Microarray and RNA-seq Data
- ComBat (2007). [Article1] [Article2] [GitHub] removing known batch effects using empirical Bayes frameworks
- svaseq (2014). [Article] [Bioconductor] [GitHub] removing batch effects with known control probes
- Seurat v2 (2018). [Home] [Article] [GitHub] [Lab] Tools for Single Cell Genomics
- fastMNN (2018). [Article] [Bioconductor] [GitHub] [Lab] batch effect correction by matching mutual nearest neighbors
- MOFA (2018). [Home] [Article] [GitHub] [pdf] Multi-Omics Factor Analysis
- LIGER (2019). [Article] [CRAN] [GitHub] [PyLiger] [Lab] Linked Inference of Genomic Experimental Relationships
- Harmony (2019). [Home] [Article] [CRAN] [GitHub] [harmonypy] [Lab] Fast, sensitive and accurate integration of single-cell data
- Scanorama (2019). [Home] [Article] [GitHub] [Lab] Panoramic stitching of single cell data
- GeoSketch (2019). [Home] [Article] [GitHub] [Lab] Geometric sketching compactly summarizes the single-cell transcriptomic landscape
- MOFA+ (2020). [Home] [Article] [Bioconductor] [GitHub] [pdf] Multi-Omics Factor Analysis v2
- MEFISTO (2020). [Home] [Article] [BioRxiv] [GitHub] [pdf] a flexible and versatile toolbox for modeling high-dimensional data when spatial or temporal dependencies between the samples are known
- Cellij (2023). [Article] [GitHub] [pdf] A Modular Factor Model Framework
-
SCENIC (2017). [Home] [GitHub] [Lab] [pySCENIC] [Tutorials] single-cell regulatory network inference and clustering
- GENIE3. [GitHub] [Bioconductor] [Vignette]
- GRNBoost. [GitHub]
- RcisTarget. [GitHub] [Bioconductor] [Vignette]
- AUCell. [GitHub] [Bioconductor] [Vignette]
-
SCENIC+ (2023). [GitHub] [Lab] [Tutorials] single-cell multiomic inference of enhancers and gene regulatory networks
-
scArches (2020). [Article] [Docs] [GitHub] [Theis Lab] [pdf] Single-cell architecture surgery, a package for reference-based analysis of single-cell data
- scVI (Lopez et al., 2018)
- trVAE (Lotfollahi et al., 2020)
- scANVI (Xu et al., 2019)
- scGen (Lotfollahi et al., 2019)
- expiMap (Lotfollahi et al., 2023)
- totalVI (Gayoso al., 2019)
- treeArches (Michielsen et al., 2022)
- SageNet (Heidari et al., 2022)
- mvTCR (Drost et al., 2022)
- scPoli (De Donno et al., 2022)
-
Mowgli (2023). [Article] [Doc] [GitHub] [pdf] Multi Omics Wasserstein inteGrative anaLysIs
- scBasset (2021). [Article] [BioRxiv] [GitHub] [Calico Labs] Sequence-based modeling of single cell ATAC-seq using convolutional neural networks
- scVI (2018). [Home] [Article] [GitHub] [Guide] [Yosef Lab] Single-cell variational inference tools
- scGen (2018). [Article] [Docs] [GitHub] [Theis Lab] [pdf] Single cell perturbation prediction
- trVAE (2019). [Article] [GitHub] [Theis Lab] Conditional out-of-distribution prediction using transfer VAE
- scANVI (2019). [Home] [Article] [BioRxiv] [GitHub] [Yosef Lab] single-cell ANnotation using Variational Inference
- VEGA (2020). [Article] [Docs] [GitHub] VAE Enhanced by Gene Annotations
- totalVI (2020). [Home] [Article] [BioRxiv] [GitHub] [Yosef Lab] Total Variational Inference
- PeakVI (2021). [Home] [Article] [BioRxiv] [GitHub] [Yosef Lab] [pdf] A Deep Generative Model for Single Cell Chromatin Accessibility Analysis
- MultiVI (2021). [Home] [Article] [BioRxiv] [GitHub] [Yosef Lab] [pdf] A deep generative model for the integration of multimodal data
- CPA (2021). [Article] [GitHub1] [GitHub2] [Theis Lab] [pdf] The Compositional Perturbation Autoencoder learns effects of perturbations at the single-cell level
- chemCPA (2022). [Article] [GitHub] [Theis Lab] [pdf] Predicting Cellular Responses to Novel Drug Perturbations at a Single-Cell Resolution
- MrVI (2022). [BioRxiv] [GitHub] [Yosef Lab] [pdf] Multi-resolution Variational Inference for quantifying sample-level heterogeneity in single-cell omics
- scGCN (2020). [Article] [BioRxiv] [GitHub] [pdf] a graph convolutional networks algorithm for knowledge transfer in single cell omics
- GLUE (2021). [Article] [BioRxiv] [GitHub] [pdf] Graph-Linked Unified Embedding
- DeepMAPS (2021). [Article] [BioRxiv] [GitHub] [pdf] Single-cell biological network inference using a heterogeneous graph transformer
- GEARS (2022). [Article] [BioRxiv] [GitHub] [Leskovec Lab] [pdf] a geometric deep learning model that predicts outcomes of novel multi-gene perturbations
- scBERT (2021). [Article] [GitHub] [Lab] [pdf] a large-scale pretrained deep language model for cell type annotation
- tGPT (2022). [Article] [BioRxiv] [HuggingFace] [Lab] [pdf] Generative pretraining from large-scale transcriptomes for single-cell deciphering
- Geneformer (2022). [Article] [Data] [HuggingFace] [Lab] [pdf] Transfer learning enables predictions in network biology
- scFormer (2022). [BioRxiv] [GitHub] [Lab] [pdf] a universal representation learning approach for single-cell data using transformers
- SATURN (2023). [BioRxiv] [GitHub] [Leskovec Lab] [pdf] Towards Universal Cell Embeddings: Integrating Single-cell RNA-seq Datasets across Species with SATURN
- xTrimoGene (2023). [BioRxiv] [Lab] [pdf] an efficient and scalable representation learner for single-cell RNA-seq data
- scGPT (2023). [BioRxiv] [GitHub] [Lab] [pdf] towards building a foundation model for single-cell multi-omics using generative AI
- scFoundation (2023). [BioRxiv] [GitHub] [Lab] [pdf] large scale foundation model on single-cell transcriptomics
- GET (2023). [BioRxiv] [GitHub] [HuggingFace] [Lab] [pdf] a foundation model of transcription across human cell types
- CellPolaris (2023). [BioRxiv] [GitHub] [pdf] Decoding Cell Fate through Generalization Transfer Learning of Gene Regulatory Networks
- GeneCompass (2023). [BioRxiv] [GitHub] [pdf] Deciphering Universal Gene Regulatory Mechanisms with Knowledge-Informed Cross-Species Foundation Model
- CellPLM (2023). [BioRxiv] [GitHub] [Lab] [pdf] Pre-training of Cell Language Model Beyond Single Cells
- GenePT (2023). [BioRxiv] [GitHub] [Lab] [pdf] GenePT: A Simple But Hard-to-Beat Foundation Model for Genes and Cells Built from ChatGPT
- UCE (2023). [BioRxiv] [GitHub] [Leskovec Lab] [pdf] Universal Cell Embeddings: A Foundation Model for Cell Biology
- A benchmark of batch-effect correction methods for single-cell RNA sequencing data. (2020) [Article] [GitHub] [pdf]
- Benchmarking atlas-level data integration in single-cell genomics. (2022) [Article] [GitHub] [pipeline] [pdf]
- Microsoft/zero-shot-scfoundation [BioRxiv] [GitHub] [pdf]
- List of awesome-deep-learning-single-cell-papers by OmicsML
- List of awesome-foundation-model-single-cell-papers by OmicsML
- Survey of single cell models by Qi Liu
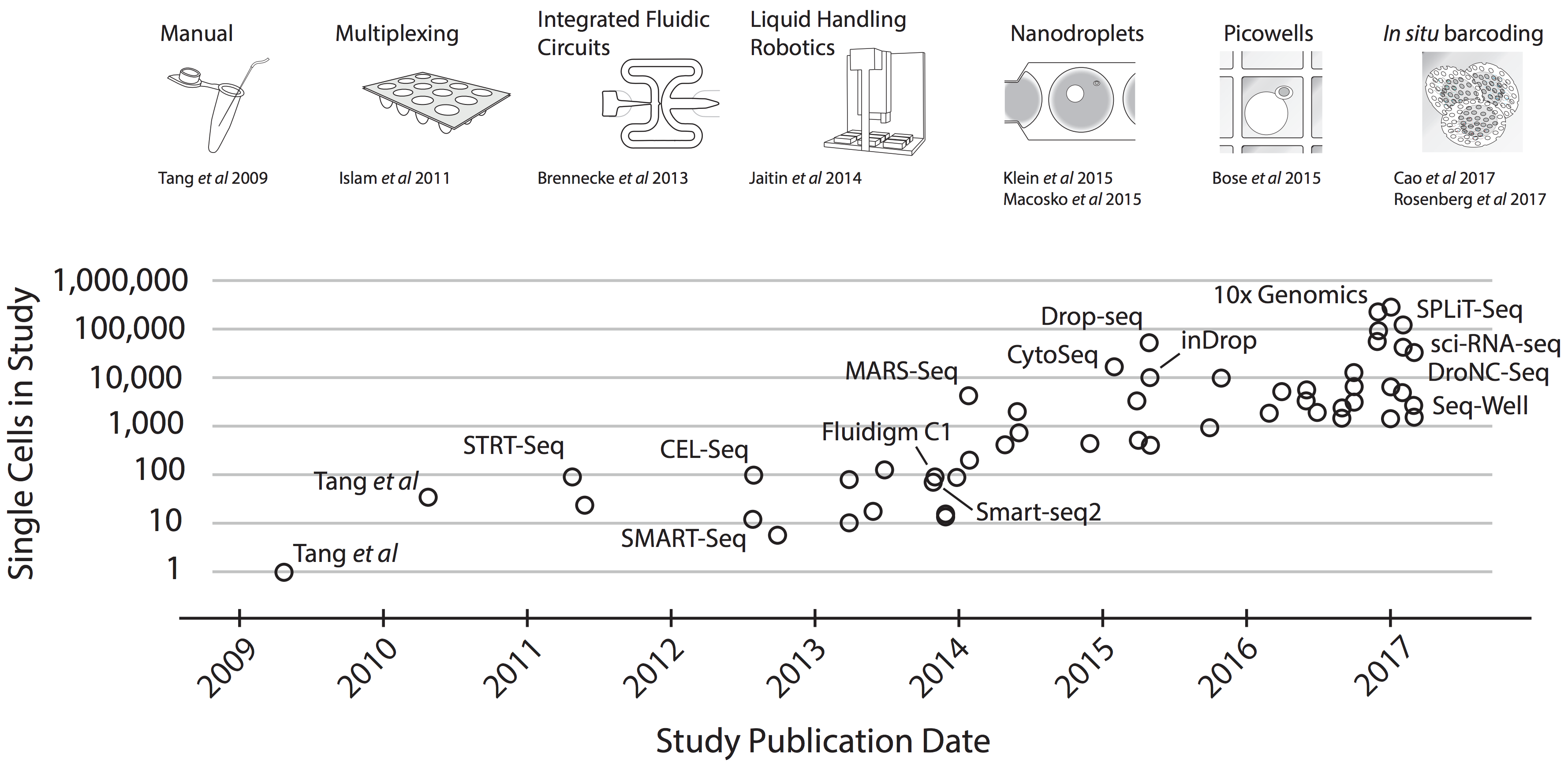
Figure 1: Scaling of scRNA-seq experiments (image from Svensson et al.)