A simple gui to help you change your molecule json files.
This is meant as an offline/intermediary solution to editing molecule json files. This will be made available via web interface in an upcoming haddock update hopefully soon. You can use this to change the PDB and/or active/passive residue.
- A working python 3 environment with pip.
- The haddock-logo and molecule images are required to be in the same map as the gui.py script.
open cmd, navigate to your map of choice. If you already have git you can skip the first step
Just copy pasta the commands below into your terminal. Say Y if it asks you a Y/n question.
pip install git
git clone https://github.com/haddocking/haddock24-json-editor
pip install -r requirements.txt
gui.py
open terminal, navigate to your map of choice. If you already have git you can skip the first step
Just copy pasta the commands below into your terminal. Say Y if it asks you a Y/n question.
sidenote: some versions of linux have issues with tkinter and/or python, while perfectly solvable this involves different steps with package managers depending on the destro. Switch to windows if you don't know how to fix these issues.
sudo apt-get install git
git clone https://github.com/haddocking/haddock24-json-editor
pip3 install -r requirements.txt
python3 gui.py
Once you have executed the script you should be presented with the following gui.
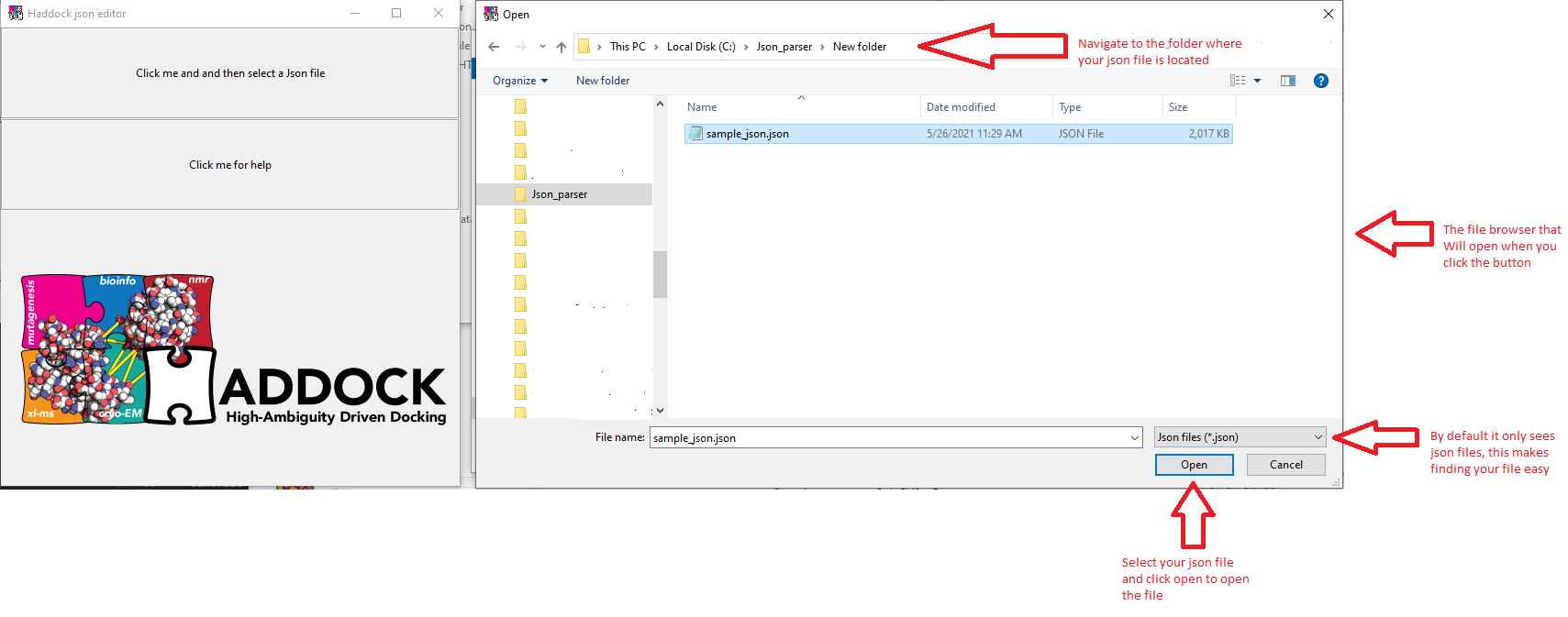
By clicking on the top button a file opener will appear and you can navigate to the folder where the json file is you wish to edit.
Upon succesfully finding and opening your json file you should be prompted with the following screen. For each molecule a button will be generated.
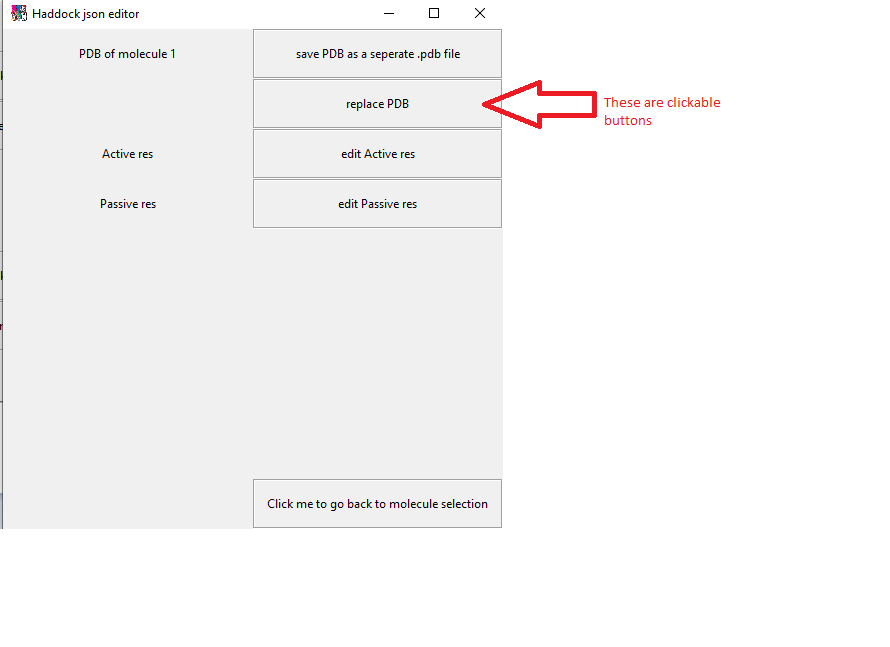
Click on the molecule of your choice you wish to edit. You will then be taken to the following screen. For now we will replace a pdb.
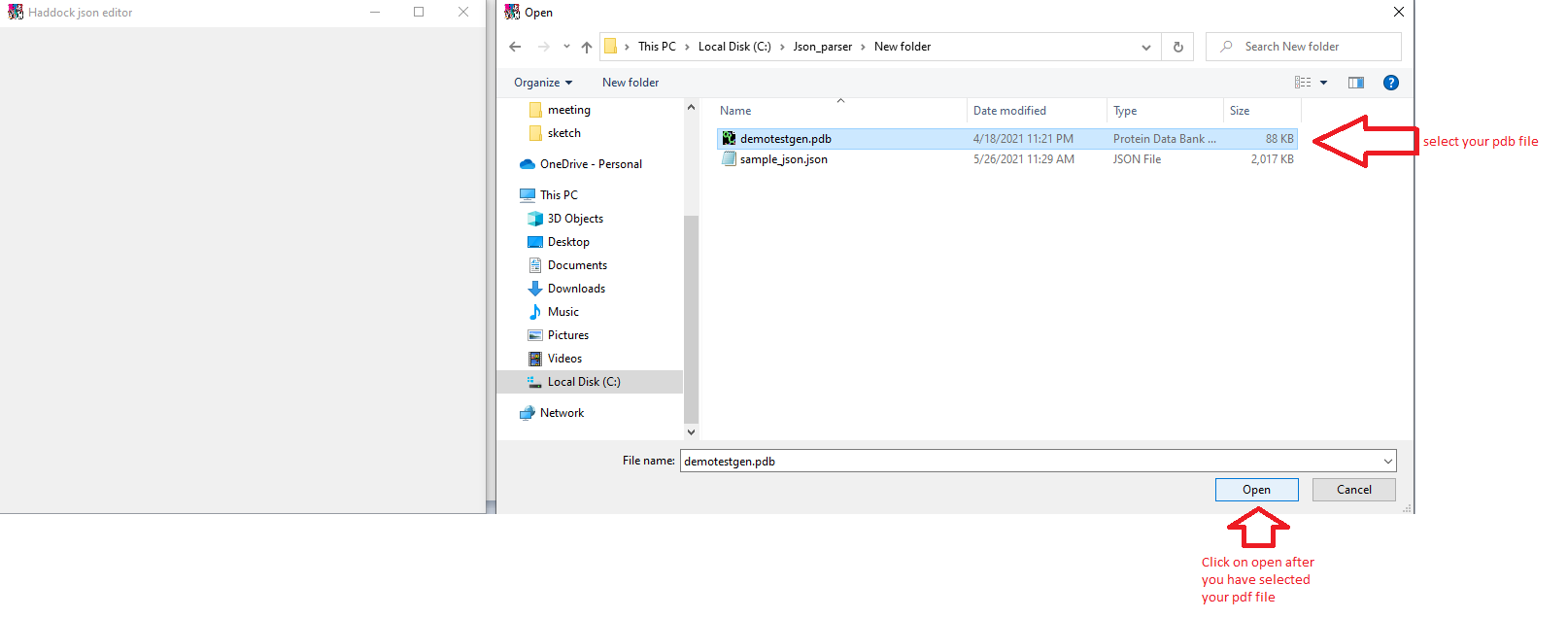
Clicking on replace pdb will open up a file browser again, use this to open up your .pdb file.
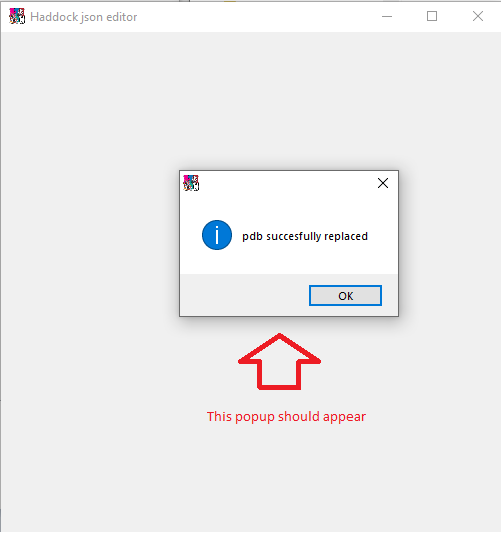
If all went well, you should recieve the following popup.
Afterwards you have replaced your pdb file you will automaticly be taken back to the molecule selection screen
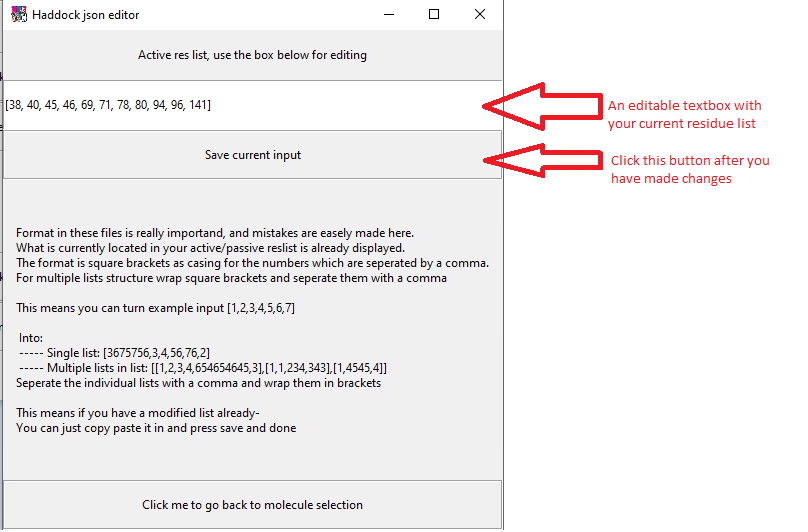
For editing a residue of a molecule, select the molecule. But this time click edit active/passive residue (they work the same). You will be presented with the following screen
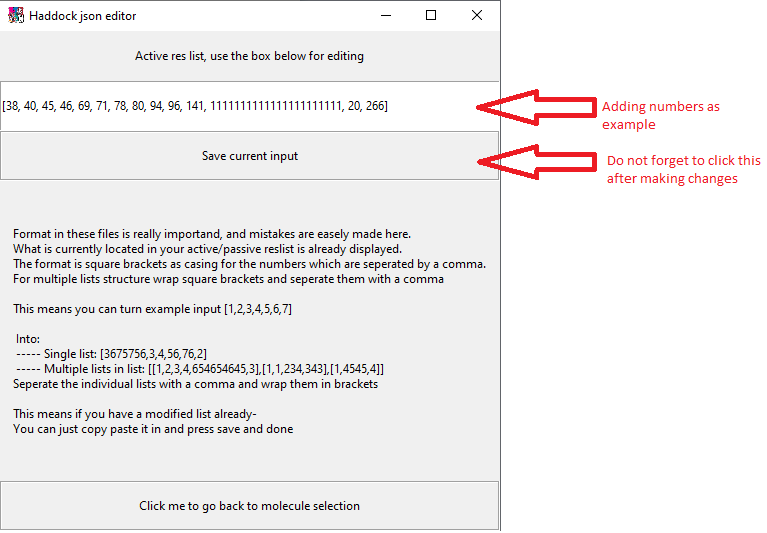
Editing a residue you can do by clicking on the text box and using your keyboard. If the residue for some reason is very large and you need to change it with another very large residue, you can use doubleclick on the textbox and then ctrl-c ctrl-v (copy paste) your new residue in.
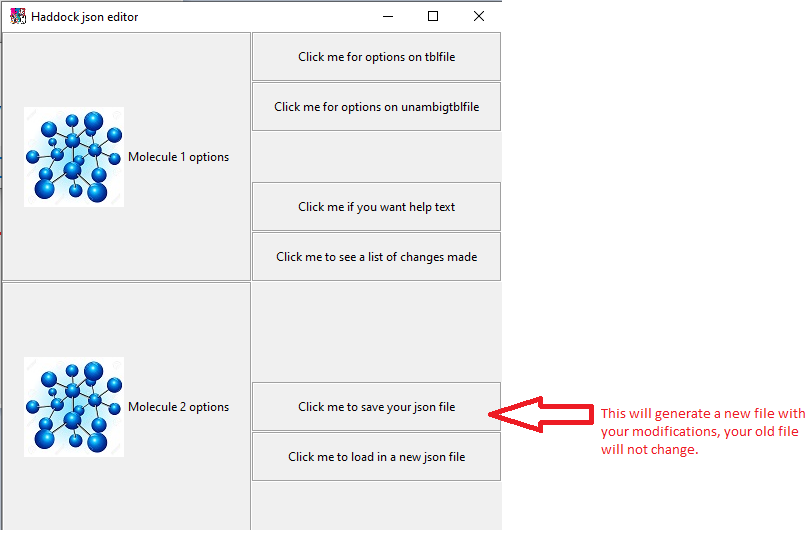
You will then be taken back to the molecule selection screen, having done our changes we now save the new file.
Save your file and done!
If you would like more, maybe different options, feel free to add requests below