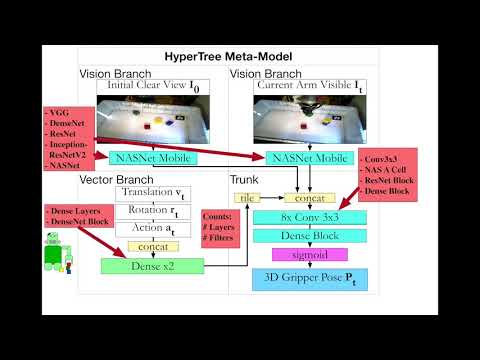
Paper: Training Frankenstein's Creature to Stack: HyperTree Architecture Search.
Authors: Andrew Hundt, Varun Jain, Chris Paxton, Gregory D. Hager.
rENAS is an extension of Efficient Neural Architecture Search via Parameter Sharing (2018) to regression problems. It is implemented in TensorFlow as a modified direct fork of the original ENAS implementation.
rENAS extends ENAS with mean squared logaritmic error (MSLE), mean squared error (MSE) loss functions and an approximate inverse of these loss functions for the reward. We have also parameterized the number of reduction and normal cells, plus replaced relu with elu, and average pooling with max pooling. We have also added support for the CoSTAR Block Stacking Dataset and the Fashion-MNIST dataset.
This material is based upon work supported by the National Science Foundation under NRI Grant Award No. 1637949.
To run rENAS search first download the CoSTAR Block Stacking Dataset, which is approximately 0.5 TB in size. Then, from the home directory run the architecture search script:
./scripts/costar_block_stacking_rotation_search_no_root.sh
This will search for architectures and print them out as it runs. You should look through these outputs to find an architecture that seems to perform particularly well, and then train that final architecture from scratch to get the best results.
To run rENAS with a final architecture determined by the search algorithm you have to specify the archiecture using a string representing the model graph. The following is an example script for using the architecture we described in our paper:
./scripts/costar_block_stacking_rotation_final.sh
The original repository for efficient neural architecture search is at https://github.com/melodyguan/enas. This is a modified version.
Includes code for CIFAR-10 image classification and Penn Tree Bank language modeling tasks.
ENAS Authors: Hieu Pham*, Melody Y. Guan*, Barret Zoph, Quoc V. Le, Jeff Dean This is not an official Google product.
The Penn Treebank dataset is included at data/ptb. Depending on the system, you may want to run the script data/ptb/process.py to create the pkl version. All hyper-parameters are specified in these scripts.
To run the ENAS search process on Penn Treebank, please use the script
./scripts/ptb_search.sh
To run ENAS with a determined architecture, you have to specify the archiecture using a string. The following is an example script for using the architecture we described in our paper.
./scripts/ptb_final.sh
A sequence of architecture for a cell with N nodes can be specified using a sequence a of 2N + 1 tokens
a[0]is a number in[0, 1, 2, 3], specifying the activation function to use at the first cell:tanh,ReLU,identity, andsigmoid.- For each
i,a[2*i]specifies a previous index anda[2*i+1]specifies the activation function at thei-th cell.
For a concrete example, the following sequence specifies the architecture we visualize in our paper
0 0 0 1 1 2 1 2 0 2 0 5 1 1 0 6 1 8 1 8 1 8 1
To run the experiments on CIFAR-10, please first download the dataset. Again, all hyper-parameters are specified in the scripts that we descibe below.
To run the ENAS experiments on the macro search space as described in our paper, please use the following scripts:
./scripts/cifar10_macro_search.sh
./scripts/cifar10_macro_final.sh
A macro architecture for a neural network with N layers consists of N parts, indexed by 1, 2, 3, ..., N. Part i consists of:
- A number in
[0, 1, 2, 3, 4, 5]that specifies the operation at layeri-th, corresponding toconv_3x3,separable_conv_3x3,conv_5x5,separable_conv_5x5,average_pooling,max_pooling. - A sequence of
i - 1numbers, each is either0or1, indicating whether a skip connection should be formed from a the corresponding past layer to the current layer.
A concrete example can be found in our script ./scripts/cifar10_macro_final.sh.
To run the ENAS experiments on the micro search space as described in our paper, please use the following scripts:
./scripts/cifar10_micro_search.sh
./scripts/cifar10_micro_final.sh
A micro cell with B + 2 blocks can be specified using B blocks, corresponding to blocks numbered 2, 3, ..., B+1, each block consists of 4 numbers
index_1, op_1, index_2, op_2
Here, index_1 and index_2 can be any previous index. op_1 and op_2 can be [0, 1, 2, 3, 4], corresponding to separable_conv_3x3, separable_conv_5x5, average_pooling, max_pooling, identity.
A micro architecture can be specified by two sequences of cells concatenated after each other, as shown in our script ./scripts/cifar10_micro_final.sh
Please consider citing both papers associated with this repository.
Please consider citing the paper introducing rENAS if you use it.
Training Frankenstein's Creature to Stack: HyperTree Architecture Search.
The rENAS citation information will be added when it becomes available, or email Andrew Hundt athundt@gmail.com for citation information.
@article{hundt2018hypertree,
author = {Andrew Hundt and
Varun Jain and
Chris Paxton and
Gregory D. Hager},
title = "{Training Frankenstein's Creature to Stack: HyperTree Architecture Search}",
journal = {ArXiv e-prints},
archivePrefix = {arXiv},
eprint = {1810.11714},
year = 2018,
month = Oct,
url = {https://arxiv.org/abs/1810.11714}
}
If you happen to use ENAS, please consider citing it.
@article{enas,
title = {Efficient Neural Architecture Search via Parameter Sharing},
author = {Pham, Hieu and
Guan, Melody Y. and
Zoph, Barret and
Le, Quoc V. and
Dean, Jeff
},
journal = {Arxiv, 1802.03268},
year = {2018}
}