This repository was archived by the owner on Oct 13, 2021. It is now read-only.
-
Notifications
You must be signed in to change notification settings - Fork 106
This repository was archived by the owner on Oct 13, 2021. It is now read-only.
Generated GRU/LSTM model with parametric seq_length size? #122
Copy link
Copy link
Closed
Description
Hello, when using the following code to convert SimpleRNN/GRU/LSTM/BI-LSTM to onnx, found that SimpleRNN/BI-LSTM are consistent to ONNX spec, while GRU/LSTM NOT.
We know that the input X of those ops should with the shape of `[seq_length, batch_size, input_size], and batch_size could be parametric.
Bi-LSTM is correct as it uses "Transpose" op, so X is 3 * N *4 as below:
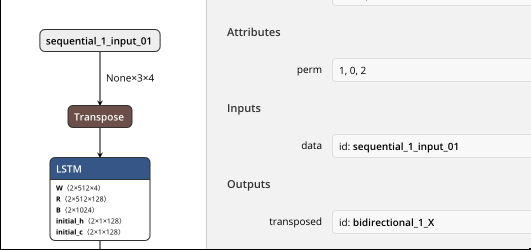
But for GRU/LSTM:
it uses "Reshape" op, so X is 6N * 1 * 4:

why setting seq_length to "6N"?
Code to generate models:
(modify SimpleRNN to LSTM/GRU will get LSTM/GRU models)
# -*- coding: utf-8 -*-
import time
import math
import numpy as np
from keras.models import Sequential
from keras.layers import SimpleRNN, GRU, LSTM
from sklearn.metrics import mean_squared_error
from keras import backend as K
import tensorflow as tf
import keras2onnx
def print_data(data):
data = np.ravel(data)
tmp = "["
for d in data:
tmp_pair = str(d) + ", "
tmp+=tmp_pair
tmp = tmp[:-2]
tmp+="]"
print(tmp)
# Fix random seed for reproducibility
np.random.seed(1)
data_dim = 4 # input_size
timesteps = 3 # seq_length
# expected input data shape: (batch_size, timesteps, data_dim)
test_input = np.random.random_sample((100, timesteps, data_dim))
test_output = np.random.random_sample((100, 128))
# Number of layer and number of neurons in each layer
num_neur = [128, 256, 128]
# Training times
epochs = 200
# Batch size
batch_size = 50
# Record time
start_cr_a_fit_net = time.time()
# Create and fit the RNN network
model = Sequential()
for i in range(len(num_neur)): # multi-layer
if len(num_neur) == 1:
model.add(SimpleRNN(num_neur[i], input_shape=(timesteps, data_dim), unroll=True))
else:
if i < len(num_neur) - 1:
model.add(SimpleRNN(num_neur[i], input_shape=(timesteps, data_dim), return_sequences=True, unroll=True))
else:
model.add(SimpleRNN(num_neur[i], input_shape=(timesteps, data_dim), unroll=True))
# Summary the structure of neural network
model.summary()
# Compile the neural network
model.compile(loss='mean_squared_error', optimizer='adam')
# Fit the LSTM network
model.fit(test_input, test_output, epochs=epochs, batch_size=batch_size, verbose=0)
end_cr_a_fit_net = time.time() - start_cr_a_fit_net
print('Running time of creating and fitting the LSTM network: %.2f Seconds' % (end_cr_a_fit_net))
print('model.inputs', model.inputs)
print('model.outputs', model.outputs)
test_input = np.random.random_sample((1, timesteps, data_dim)).astype(np.float32)
test_output = model.predict(test_input)
onnx_model = keras2onnx.convert_keras(model, model.name)
import onnx
onnx_filename = "keras_rnn1.onnx"
onnx.save_model(onnx_model, onnx_filename)
K.clear_session()
#####################
# verify
#####################
import onnxruntime as rt
sess = rt.InferenceSession(onnx_filename)
input_name_X = sess.get_inputs()[0].name
label_name_Y = sess.get_outputs()[0].name
pred_rst = sess.run([label_name_Y], {input_name_X: test_input})
Metadata
Metadata
Assignees
Labels
No labels