-
Notifications
You must be signed in to change notification settings - Fork 189
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
we need a data catalog #39
Comments
|
Perhaps just some yaml file on the network file system that we refer against as well as some tiny library that reads that file? Also cc'ing @seibert , who thinks about these things within Anaconda Inc. @seibert these tend to be glob-string paths pointing to NetCDF files. All machines have uniform access to them, so there is no need for remote data access. Modeling data types would be difficult due to the complexity of the underlying files. |
|
I had a quick email exchange with @jhamman, who pointed me to this issue. Anaconda has a new open source project to provide some functionality for building data catalogs Intake. This week, I have been investigating Intake for another project and started a plugin for NetCDF. So far, |
|
Sounds very promising @mmccarty! The cataloging features of Intake could definitely solve some of the problems we are contemplating. One complication for Pangeo is that most of our cloud-based data is actually stored in zarr format, not netCDF. So my recommendation would be to broaden the scope beyond netCDF to include any format that can be read by xarray. In the docs you pointed to, it says
There is a strong analogy between pandas and xarray. Pandas.Dataframe is the fundamental python data structure. The pandas data itself can come from csv, hdf, xls, many different file formats. Likewise, xarray data can come from netcdf, hdf, geotiff, zarr, etc. What would be great is to have an xarray data catalog that is agnostic to the underlying file format. We have also discussed Quilt in #162, which I guess aims to solve a similar problem |
|
@rabernat My initial approach was to add an xarray container to Intake, however it felt a bit awkward (like this was a lower level of abstraction that we needed since xarray already provides much of the dask and file IO functionality on it's own). Also, the xarray container needed a slightly different interface to support chunks in N-dimensions. So, we (@seibert @martindurant and I) then decided to attempt to implement a plugin "the harder way", which is the approach in the netCDF plugin. Options we are considering:
|
|
It would be nice to separate netCDF from xarray in this work. There are multiple libraries for loading netCDF files into numpy/dask arrays. From our point of view we would like to be able to add support to iris. |
|
Sounds like a case for option 1 (which I favor) plus an iris Intake container in the future (this PR shows the container interface thus far). Then plugins can be built for netCDF using either containers. |
|
There would already multiple methods we would want to load xarrays - I don't know iris, but netCDF/hdf5 limited local files (or NFS, FUSE) and zarr for data anywhere. Also, there are other data formats out there that are like this such as minc (a particular netcdf flavour), dicom, fits; which all have heterogeneous data types of ndarrays (and maybe tables) and copious metadata with coordinate information. |
|
@mmccarty: let me reiterate that I am very enthusiastic this. A simple data catalog would really accelerate Pangeo. Right now we have a solid computational platform, but the platform is useless without data. So far, we have mostly been working with datasets that we already know about, loading them manually. It sounds like Intake could really help us grow a general purpose, flexible data catalog. I repeat my comment above that it is very important for us to be able to load xarray from multiple different backends. On pangeo.pydata.org, most of the data is stored in zarr format in google cloud storage. Here is what I imagine the Does this help you? Please let me know what other input you need to guide the design of xarray / intake integration. Perhaps we should open an issue specifically for that. p.s. Could you clarify what you mean by "option 1"? |
|
@rabernat , that is exactly what we envisage. The decision was whether each plugin would be written "from scratch" and implement all methods or whether we can have a "xarray" container type that is common to many. In fact, since xarray already has several loader methods, we could also have a combined plugin for all, where the entry would look something like |
|
Sounds like are on the same page! Please let us know how we can help. |
|
I think that a useful deliverable here would be a pull request that both changed the Docker image in this repository (see gce/notebook/Dockerfile) and the attached notebooks (see gce/notebook/examples/*.ipynb) that replaced current mechanisms to create XArray datasets from Zarr on GCS that look like this: import gcsfs
import xarray as xr
gcsmap = gcsfs.mapping.GCSMap('pangeo-data/dataset-duacs-rep-global-merged-allsat-phy-l4-v3-alt')
ds = xr.open_zarr(gcsmap)with something that looks like this import intake
ds = intake.open('dataset-....')Presumably this would require intake to grow appropriately. It would also require the addition of an intake config file in the docker image that specified all of the dataset locations pointed to by the various notebooks. |
|
Hi, my name is Paul Branson, I am a coastal scientist from Perth, Australia and have been lurking around for a while to learn about this awesome project! From reading your recent discussions about CMIP6 (#179) and this thread I thought up a possible use case as a solution to a current problem my research group has for a distributed information system. The idea would be to setup a Kubernetes cloud federation (https://medium.com/google-cloud/experimenting-with-cross-cloud-kubernetes-cluster-federation-dfa99f913d54) that joins up multiple clouds each that is physically located with the data. Then it might be possible to write an xarray driver for INTAKE that natively starts a dask KubeCluster() with workers+backend on the appropriate pods(?) and a dataset specific yaml. I think INGRESS could be used to effectively manage the dataset namespacing somewhat transparently (https://medium.com/google-cloud/kubernetes-nodeport-vs-loadbalancer-vs-ingress-when-should-i-use-what-922f010849e0). Do you think that could be a reasonable approach (for co-operating organisations) to provide a simple platform for data analysis across distributed datasets? Possibly a little off topic sorry! |
|
The latest on plan intake-xarray. |
Specifically, we will provide thin wrappers for all the xarray.open-* functions, so from pangeo's point of view, you would make a catalogue by encoding the arguments currently in the notebooks into each YAML entry for the catalogue file. (I don't think there is any plan for intake to manage the execution environment) |
|
This sounds great! Let us know when you have a minimal working prototype we can try on pangeo.pydata.org! |
|
Work coming here: intake/intake-xarray#7 @rabernat |
|
I made a quick pre-release, so you can try it out with (but expect some rough edges!) |
|
@martindurant I suspect that people here don't have experience using
intake. You might have to take a couple extra steps and show an example of
its use. I recommend trying to modify one of the standard notebooks in the
examples directory if you have the time.
…On Fri, Apr 27, 2018 at 7:26 PM, Martin Durant ***@***.***> wrote:
I made a quick pre-release, so you can try it out with
conda install -c intake intake-xarray
(but expect some rough edges!)
—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub
<#39 (comment)>,
or mute the thread
<https://github.com/notifications/unsubscribe-auth/AASszH6qcdX4nOsivNgSC1T8mViZktW8ks5ts6kVgaJpZM4QfbC->
.
|
|
To be clear I'm suggesting that intake folks create a PR that modifies our
docker image to both include the necessary files for intake to work, and
also includes modified versions of the example notebooks bundled with the
image.
The relevant directory for the pangeo docker image is here:
https://github.com/pangeo-data/pangeo/tree/master/gce/notebook/
On Sat, Apr 28, 2018 at 2:05 AM, Matthew Rocklin <mrocklin@anaconda.com>
wrote:
… @martindurant I suspect that people here don't have experience using
intake. You might have to take a couple extra steps and show an example of
its use. I recommend trying to modify one of the standard notebooks in the
examples directory if you have the time.
On Fri, Apr 27, 2018 at 7:26 PM, Martin Durant ***@***.***>
wrote:
> I made a quick pre-release, so you can try it out with
>
> conda install -c intake intake-xarray
>
> (but expect some rough edges!)
>
> —
> You are receiving this because you commented.
> Reply to this email directly, view it on GitHub
> <#39 (comment)>,
> or mute the thread
> <https://github.com/notifications/unsubscribe-auth/AASszH6qcdX4nOsivNgSC1T8mViZktW8ks5ts6kVgaJpZM4QfbC->
> .
>
|
|
Agreed, @mrocklin . First, intake-xarray should have a minimum of documentation, but that is exactly the plan. |
|
Just want to reiterate my enthusiasm for these developments! Can't wait to see a working example of |
|
I just came across this jupyterlab extension: https://github.com/eWaterCycle/jupyterlab_thredds
This seems really cool! Something similar could probably be developed for intake. |
|
We could certainly do something like that. I'm not certain how the initial "catalogue location" would be populated (file-browser, list of recent locations, set of builtin cats?), but listing entries and their brief details, and then injecting load commands sounds very worthwhile. I'll raise the issue with Intake. |
|
Unless you missed it, @rabernat , the latest release of dask, now on the pangeo cluster, does not include my fix for HTTP downloading of the catalogue (github's headers give incorrect information about the file size). |
|
@martindurant - I'm not sure I understand your comment. In the dockerfile of #231, you specified I get the impression from your comment that intake is still not working properly on the pangeo cluster. What went wrong? Your fix for the HTTP downloading was not yet merged when 0.17.5 was release? What do we need to do to fix it at this point? |
|
Specifically, my fix for HTTP was merged into master in time, but the 0.17.5 release was not based on master, but to do with quick fixes for the pandas release that also just happened - so my fix is not in the release, and the (commented) lines for intake in the notebooks don't currently work :| @mrocklin suggested that he would want to return to installing some packages from master, but that of course comes with its own problems of things changing between image rebuilds. |
|
It looks like Matt is working on pointing us to a consistent set of commits for dask-related packages over in pangeo-data/helm-chart#29. Can you inspect that PR and verify whether it includes the fixes necessary for intake to work? |
|
Yes, this line includes the code: |
|
This issue has been automatically marked as stale because it has not had recent activity. It will be closed if no further activity occurs. Thank you for your contributions. |
|
This issue has been automatically closed because it had not seen recent activity. The issue can always be reopened at a later date. |
|
A more general and scalable way to create a catalog for Pangeo would be to create ISO metadata records for each Pangeo dataset and harvest them into pycsw, which would then support catalog search using CSW, the Open Geospatial Consortium Catalog Service for the Web. Users could then search for Pangeo datasets across multiple pangeo deployments, and also would provide a mechanism for pangeo datasets to be discovered on sites like http://data.gov and http://data.gov.uk. We use this approach in the Integrated Ocean Observing System (IOOS), supporting both user exploration and automated workflows. See this example notebook that finds coastal wave data from multiple providers based on a specified bounding box and time range. In IOOS, data providers serve data primarily via THREDDS and ERDDAP, both of which automatically generate ISO metadata records from NetCDF datasets with CF Conventions. In Pangeo, we don't yet have a way to automatically generate ISO metadata records from the CF Compliant NetCDF or Zarr datasets, but we could solve this by developing python code to generate ISO metadata records directly from Xarray or Iris. The logic of converting NetCDF attributes to ISO is contained in this XSL. We won't solve this tomorrow, but it might be a nice little project for the roadmap! |
|
@rsignell-usgs , Do you see a role for Intake in this supra-cataloging that you describe? Should it be able to interact with CSW, or should it have a role in extracting appropriate metadata from netCDF/zarr files? |
|
I don't know much about Intake, but I guess Intake catalogs could be constructed from the responses to CSW queries if that is useful. |
|
Finally announcing intake officially: https://www.anaconda.com/blog/developer-blog/intake-taking-the-pain-out-of-data-access/ |
|
This issue has been automatically marked as stale because it has not had recent activity. It will be closed if no further activity occurs. Thank you for your contributions. |
|
This issue has been automatically closed because it had not seen recent activity. The issue can always be reopened at a later date. |
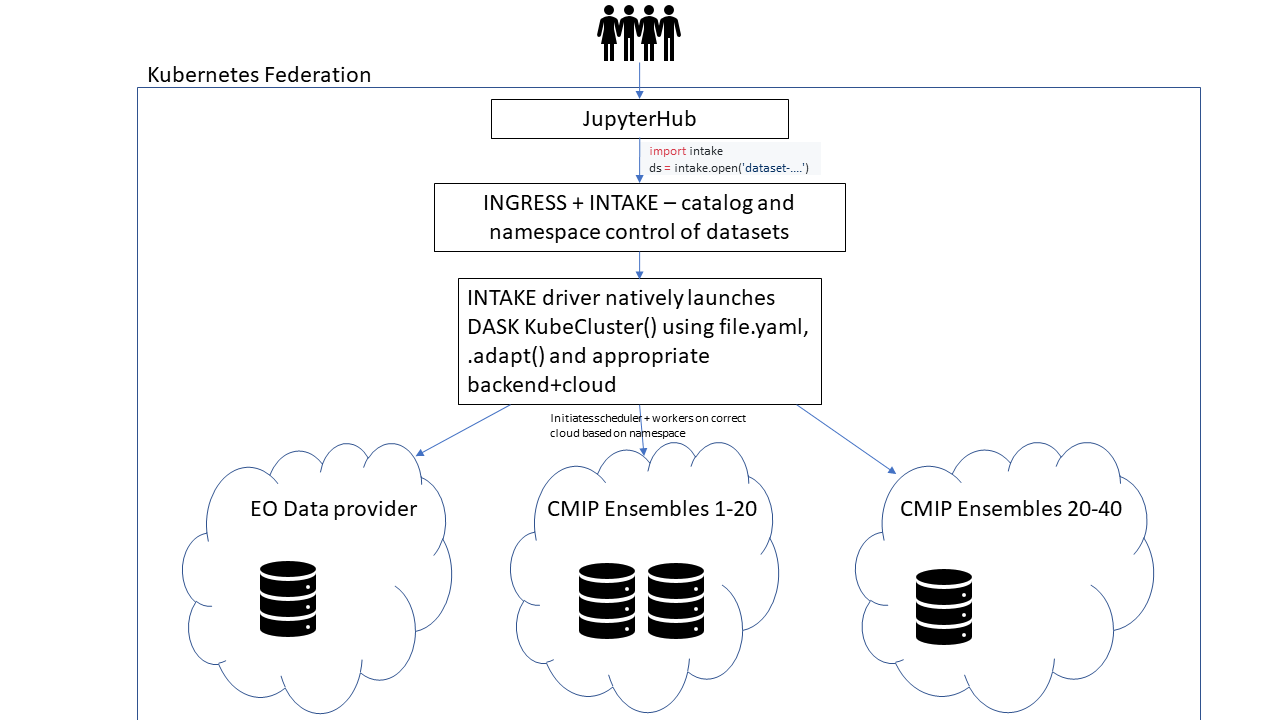
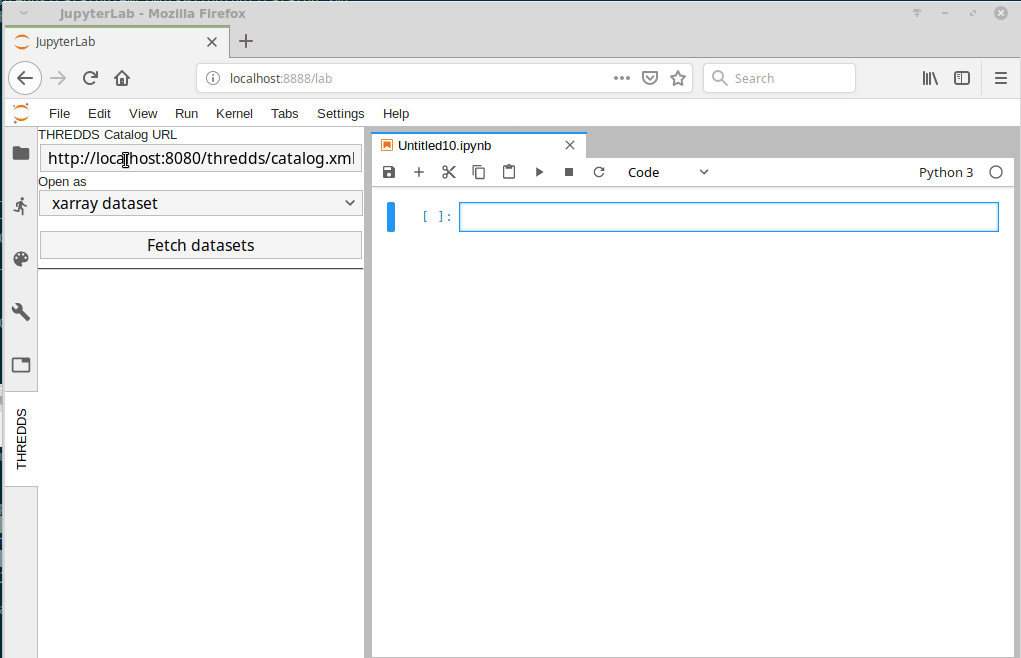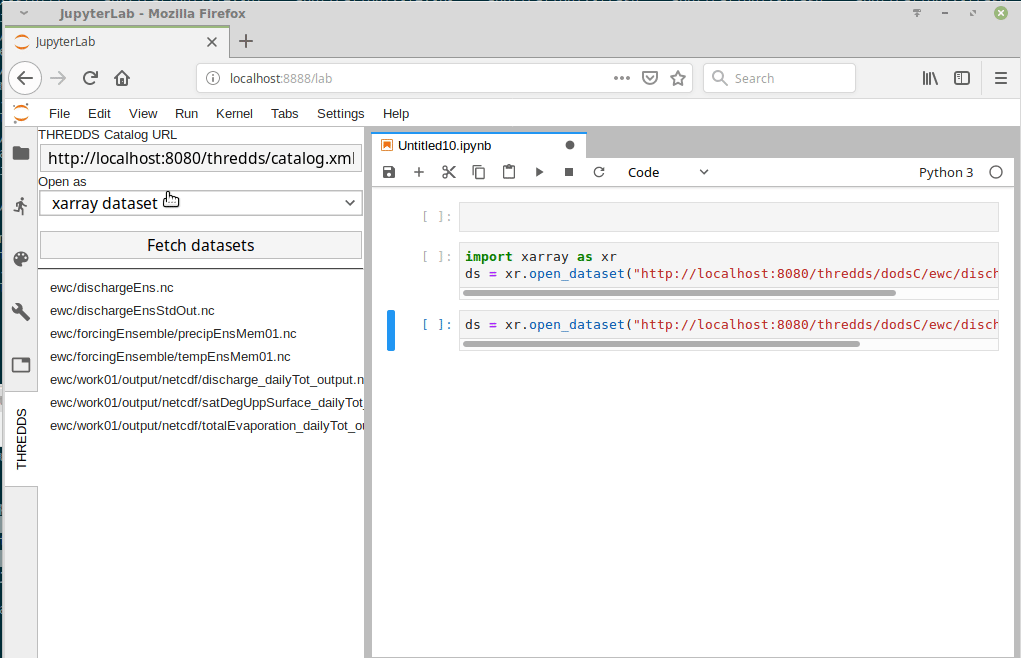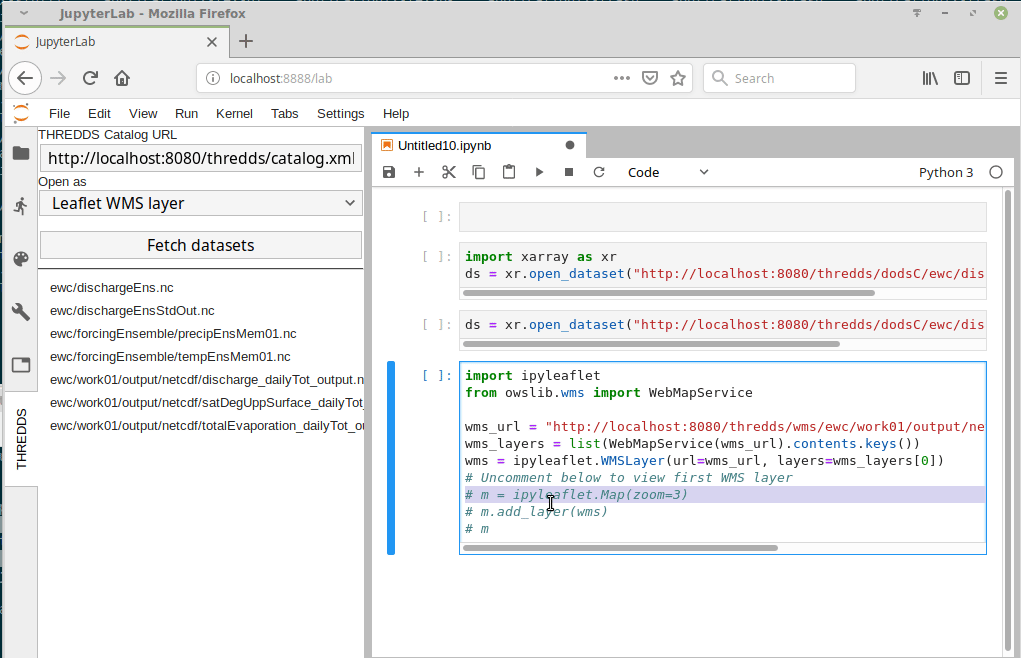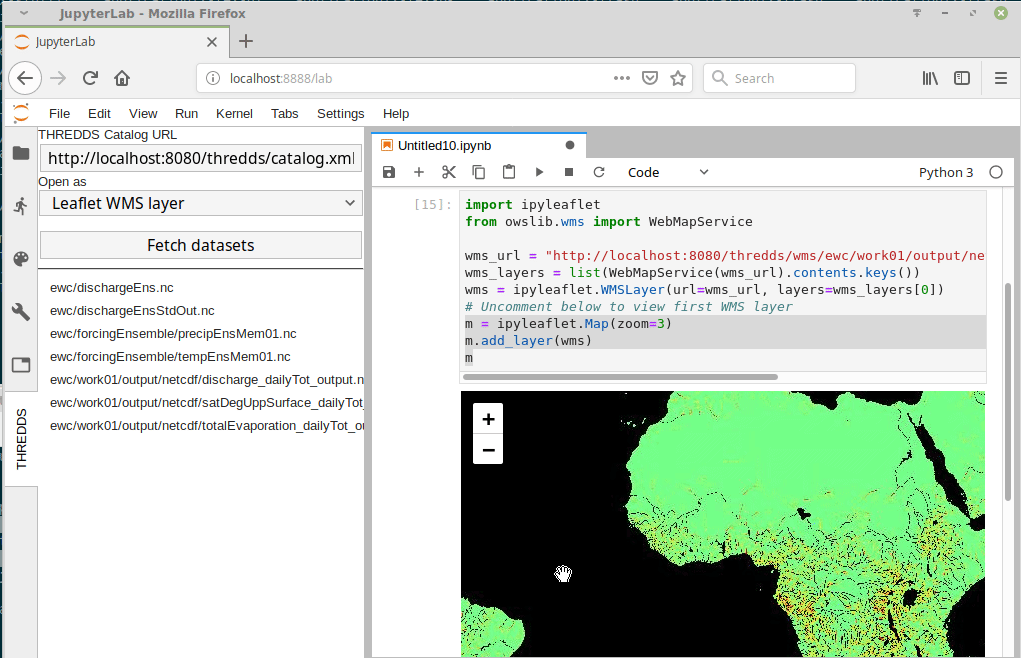
At this point we have multiple datasets on multiple different systems.
It would be great to have some kind of catalog interface to avoid having to manually change the path names every time. For example.
The text was updated successfully, but these errors were encountered: