-
-
Notifications
You must be signed in to change notification settings - Fork 26.2k
[MRG] new K-means implementation for improved performances #11950
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
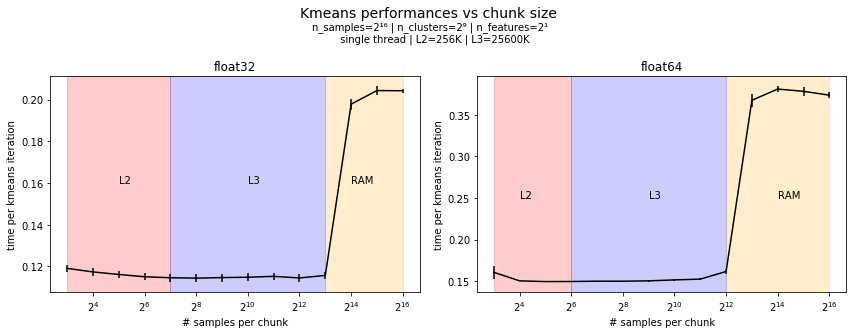
Here comes some benchmarks showing the importance of choosing the right chunk size. The following plot shows the time of a single kmeans iteration for several chunk sizes. The chunk size is the number of samples per chunk. This correspond to a memory allocation of size If the chunk size is too small (<2⁷ here) the overhead of
Note however that this is on single thread and that it will have to be adjusted for multi-threads. I'm running benchmarks for that and I'll put them here. |
|
Nice work! though I've not yet looked at the code... All the CI say no, btw. I would think cases with n_clusters < 2**3 are very common. Is there a good reason not to benchmark those smaller numbers? |
There are still a lot of holes in my code :) Especially I didn't implement the sparse case.
Absolutely not. I'll run them. Edit: results are quite similar. For instance the benchmark below is for 2 clusters in 2 dimensions. I also added the same benchmarks with data stored as np.float64. |
|
Another benefit of this implementation is that the memory usage should be limited. In particular that should also probably fix #9943 (I have not checked). |
|
The intel compiler is impressive. I wonder if we would observe similar results on the rest of the scikit-learn cython code base. If it's the case it's worth considering this as the compiler used to generate the scikit-learn wheels. |
|
@jeremiedbb is your benchmark of compilers in #11950 (comment) running on a single thread? The implementation of openmp in clang/llvm should be similar to the one in icc as it was contributed by intel. |
|
@jeremiedbb BTW congrats on the quality of the reporting of your benchmarks and the analysis in terms of CPU cache size. This is really enlightening and appreciated. |
|
Also note that when @jeremiedbb mentions the "intel implementation" of K-means, it's the one from the intel distribution of scikit-learn. In particular the K-means C++ implementation is provided by DAAL: https://github.com/intel/daal/tree/daal_2018_update3/algorithms/kernel/kmeans |
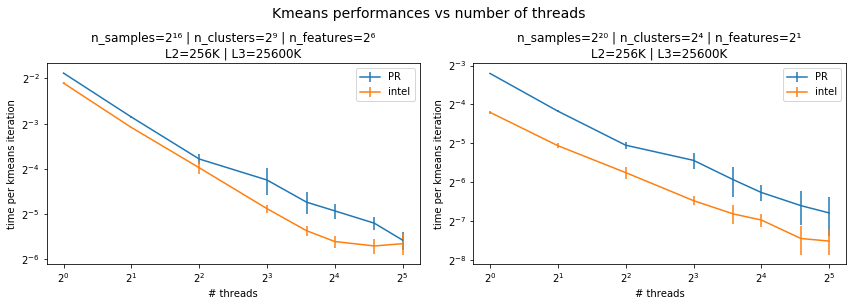
yes it's on single thread. Here is a benchmark on how it behaves on multi-threads (for 2 problems with different number of clusters and features), compared to the intel implementation. |
|
I have a hard time reading times in power of 2. Please use regular (decimal) tickers and a log scale in matplotlib instead. The scalability looks good though :) |
|
Also maybe use 10x the number of samples to get more interesting running times. We are really not interested in optimizing k-means in the subsecond regime :) |
I agree but for this benchmark, we are not interested in the absolute time. I put base 2 because I find it easier to see if time is divided by 2 when the number of threads is doubled |
|
Quick note about numerical precision issues K-means being based on euclidean metric, it's impacted by #9354. Although we don't use However, the absolute values of the distances are irrelevant for this algorithm. What matters is their ordering. Therefore this numerical precision should have a minor effect on the clustering. I've made some tests and they seem to confirm that intuition. Below is an example of the clustering using the fast and the exact distance computation (data stored on float32). For this example, the final inertiae differ by about 1%. I made other tests with various kinds of clusters, scales, numbers, dimensions, and I haven't found a situation where this numerical precision issue disturbs the behavior of the algorithm. Therefore I suggest that we keep this fast method for computing the pairwise distances in K-means. There is still a visible effect. Although the clustering is fine, the absolute distances might not be and the inertia could be wrongly evaluated. This is because the inertia is computed incrementally by the distances of each sample to it's closest center using the fast method. The solution is to compute the inertia using the stable method, once the labels are updated: |
|
Very nice work on this in general!
Well the thing is as illustrated in #9354 (comment) for vectors closer than some threshold, the distance may be 0.0 while it shouldn't be, and then your ordering will also not be correct. But I agree that as far as this PR goes, one might as well ignore it, it's a more general issue... |
|
Yes but for a sample to be wrongly labeled due to that issue, it means that the distance between this sample and say 2 centers are very close. Hence you can't say that this sample should belong to one cluster or the other. Moreover, inertia should be almost the same in both case, and you end up in 2 different local minima, with approx the same score. |
|
Can I ask how much time the total time until some sort of convergence is on these benchmarks? |
|
The y axis on my plots shows the time for a single iteration of k-means. You get the same speed up factor for the full Let's say for example you have 100000 samples, 100 clusters, 10 features. One iteration takes ~25ms. If you have around 100 iterations until convergence ( Now if you have 10x more samples it's 25s vs 50s+. Moreover, the parallelization is now at the loop over samples level, which allows the possibility to use all cores. With the current parallelization being at the init level (which default is 10), you can only use For a concrete example, let's take the example of color quantization in sklearn, but fitting the whole image, not a subsample. On master, it takes 164s, vs 27s with this implementation (on single thread). |
|
Thanks for the explanation. |
|
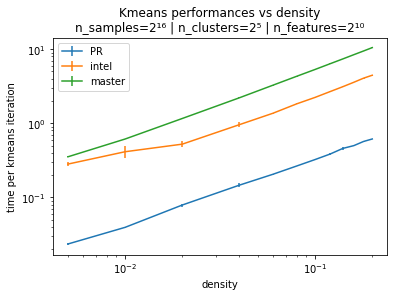
Below are some benchmarks for the sparse case. This one is a benchmark of the performances versus the number of features, for 8 clusters on the left and 128 on the right. This is on single thread. and in terms of speedup Here is a benchmark of the performances versus the density of the data sparse matrix. and in terms of speedup |
|
One way to explain the difference with intel, is that intel is using sparse matrix matrix multiplications from MKL to compute the distance while the Cython code of @jeremiedbb is computing the distances naively but leveraging the sparsity. Apparently for this problem, not using matrix matrix multiplications is the best strategy. |
|
Here is a benchmark for multithread scalability with sparse data. I'm not sure why the performances decrease at 32 threads. It might be that the gil acquisition for the reduction begins to cause a noticeable overhead, but it's more a guess than an explanation. Edit: I don't know what is the reason of the perf decrease in the previous benchmark, but it might be related to some unknown about the machine I'm running them on (virtual cpus). Because I ran it again and the results are as expected. I would expect this kind of difference if one machine use hyperthreading (i.e. 16 actually physical cores) and the other has 32 physical cores. Anyway, scalability looks good. Maybe sometimes not smart to use all available cores. |
|
I have a more formal justification about why numerical precision issues from euclidean distances don't impact k-means, or at least this new implementation. First, let's see where this issue comes from. Let x be a sample point and c be a center. Let's suppose that x and c are close (otherwise, x would be eventually assigned to a closer center and bad computation of the distance between x and c would be irrelevant), that is Then, due to precision limit, an error of order The thing which save us here is that we do not compute all the terms in the distance expansion. When we need to find the closest center for a given sample minc(d(x,c)) = minc||x-c||² = minc(||x||² + ||c||² - 2x.c) we don't need to compute Here an error of order |
|
@NicolasHug I thought I could delete comments that were duplicated as answers in previous review and comments in new review but in tunred out that it deleted both :/ |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM
Thnanks for the great work @jeremiedbb
|
@NicolasHug I think I addressed most of your comments (still remains the testing of some private utility functions). I marked them as resolved when I applied your comments as is, and left open the others. You can mark those as resolved if you are satisfied by my answers. |
|
I tried to resolve my comments yesterday but github was giving me errors so I'll leave it like that, they're all minor or addressed anyway. I haven't merged because I wasn't quite sure whether you wanted to push a few last touches. Feel free to self merge! |
|
I did another pass after @NicolasHug's review comments have been addressed. It still LGTM. Great work, merging! |
|
i noticed in the scikit-learn v0.23 release notes that the KMeans input parameter for n_jobs is deprecated and will be removed in v0.25. all that is mentioned is that the parallelization is being done differently now (previously it was done via i use the kmeans and i also use the n_jobs param and it does speed things up. my question is. does the new v0.25 version without n_jobs still parallelize yes or no? is the parallelize just automatic (no need for user because it knows what is best)? |
n_jobs can still be used in 0.23 and 0.24. However, if you leave the default value, kmeans will now use as many cores as possible.
from 0.25, it will still use all cores. There are tools to modify the number of core used described in the user guide, such as the |
|
thanks for the speedy reply and the clarification. glad to hear the parallel processing is still there and just changing the implementation of how the parallelization is done. for my own edification: if you are parallelizing at the openmp/c++ level i guess your code is already c++ and therefore better to do parallel processing at c++/low-level as opposed to the old way of parallelizing at the higher-level python level with joblib? |
|
here's a blog post I wrote about that which describes the new optimizations and the change in the parallel scheme. |













This is work in progress toward an improved implementation of K-means, performance-wise. It's an attempt to implement some of the propositions in #10744.
Currently this is only done for the lloyd algorithm.
The main difference with the current implementation is running the steps of the algorithm over data chunks, allowing better cache optimization and parallelization.
Although this is still WIP and there's a lot more work to do, any comment is appreciated.
Currently there are 2 implementations of lloyd algorithm, triggered by the
precompute_distancesparameter. When False, the pairwise distances between the data points and the centers are computed in a cython loop, using level 1 BLAS functions. This is not efficient. When True, the pairwise distances are precomputed before the cython loop, using level 3 BLAS functions through numpy.dot. This is very efficient. However the obtained pairwise distances matrix can be quite large and does not fit in the cpu cache which may cause a big overhead.The proposed implementation tries to solve both issues. The pairwise distance matrix is precomputed using level 3 BLAS functions, one data chunk at a time. This would result in simpler implementation: no need for 2 different implementations.
Also it may also allow a better parallelization, at the chunks loop level (although this need more test right now).
This first implementation shows improvement, on single thread, by a factor of 2 to 8 versus master (I'll post some benchmarks soon). However it is still slower than the intel implementation by a factor of 2, and I need to investigate further.
There is still a long todo list
publish benchmarks on single and mutli thread.
find good chunk size is important for the performances
add sample weight
support sparse
clean now unnecessary code
investigate the elkan algorithm
investigate the centers initialization (kmeans++)(will do in separate PR)Fixes #9943, fixes #10744